Methods for screening and using crispr/cas9 guidance RNA sequence from HIV provirus genome
a genome and genome technology, applied in the direction of viruses/bacteriophages, biochemistry apparatus and processes, peptides/protein ingredients, etc., can solve the problems of serious dysregulation of neuronal function and homeostasis, astrocytes' ability to maintain homeostasis, and aids remains incurabl
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Establishment of Stable Cas9 Expressing Astrocyte Cell Line
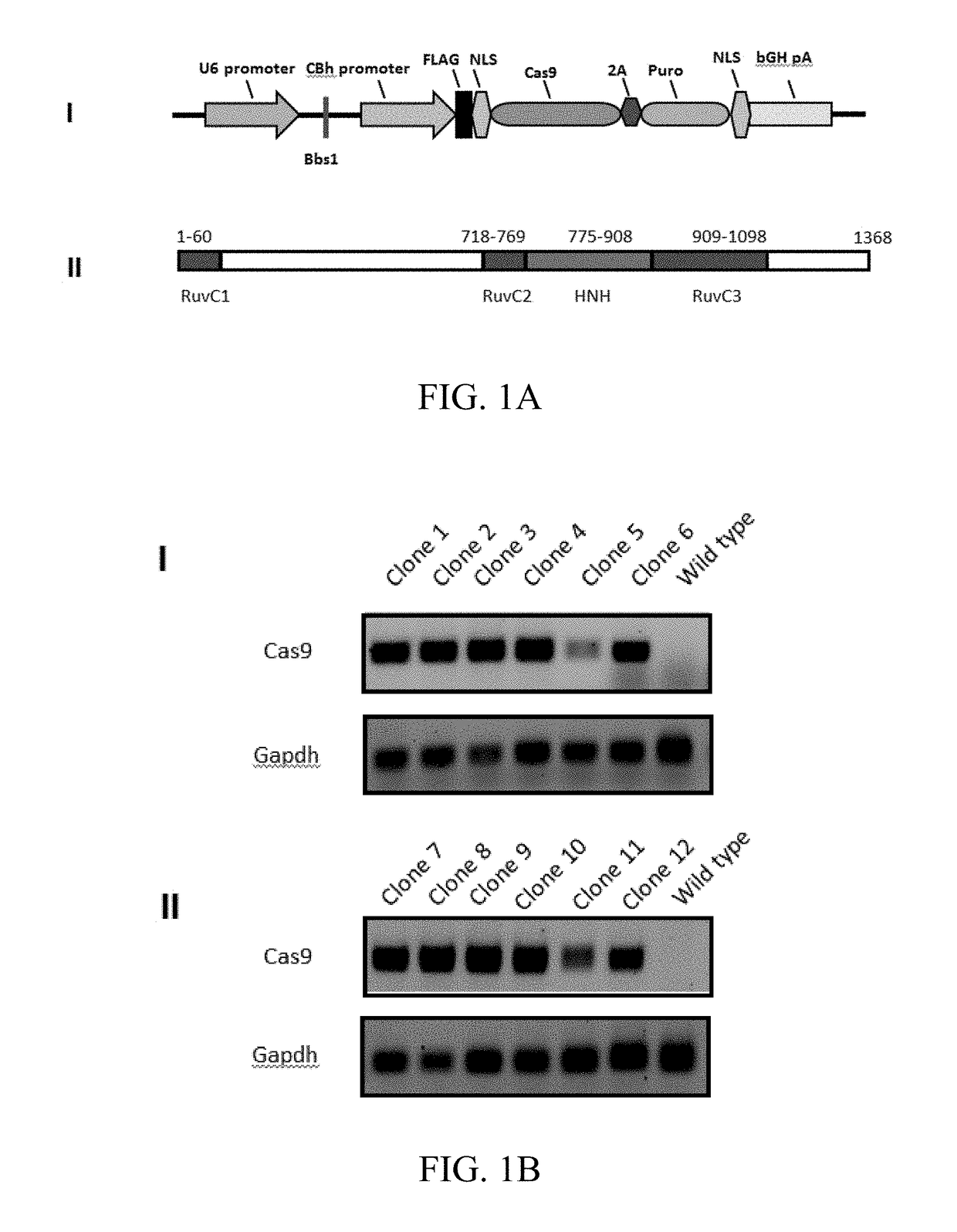
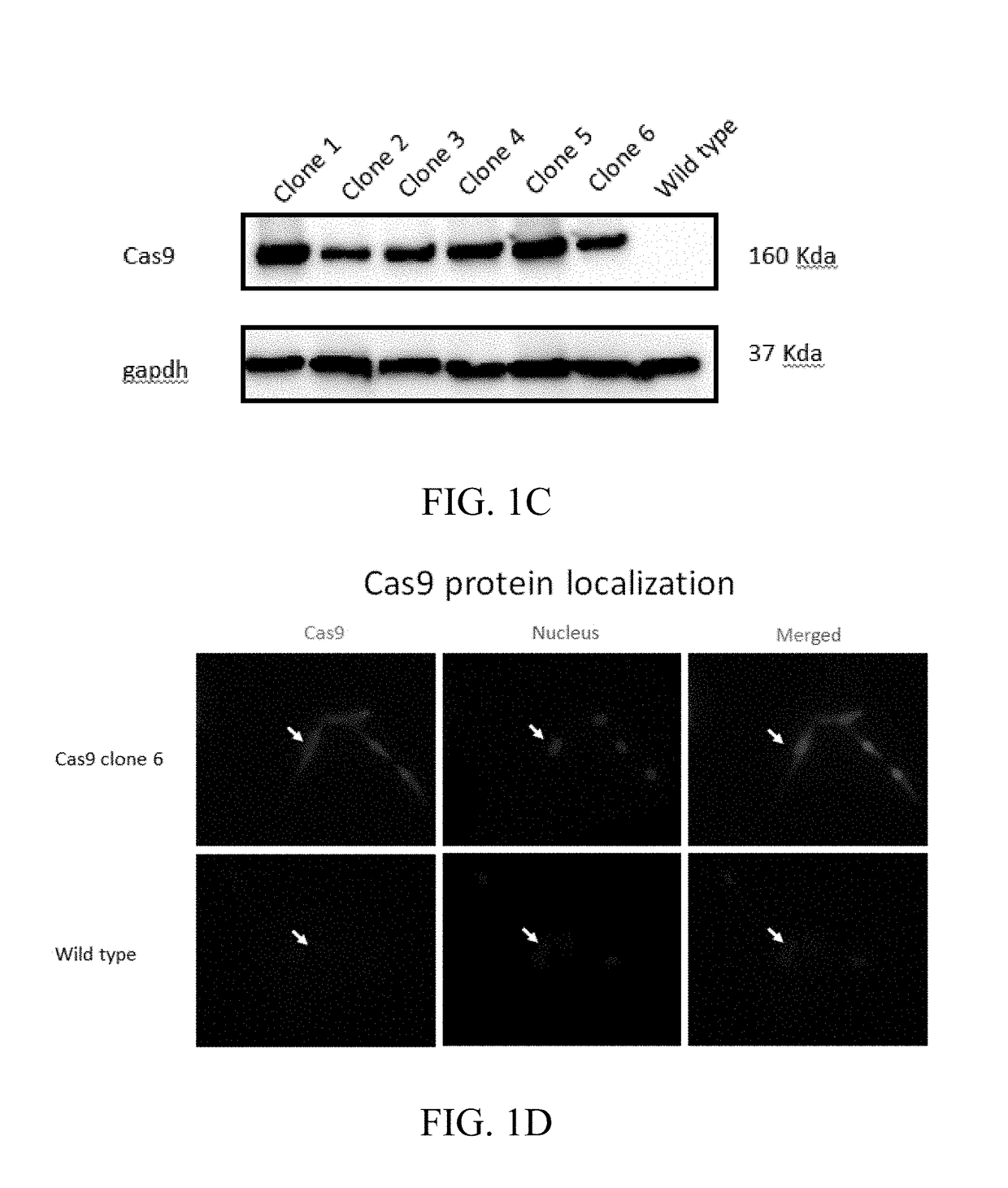
[0096]A genetically engineered astrocyte cell line stably expressing Cas9 was produced. An astrocyte cell line was transfected with intact or linearized Cas9 plasmid pX 459 and cell clones were selected for Cas9 plasmid integrated based on puromycin resistance. By testing the death curve of astrocyte under the puromycin resistance, the concentration of 0.31 μg / ml was chosen for selection. More positive cell clones were obtained from the linearized plasmid group compared to intact plasmid. The cell clones were expanded and genomic DNA was extracted for a PCR screen of Cas9 gene integration. Over ten cell clones provided a specific PCR product band, which was absent from the parent cell line (FIGS. 1B-1C). After the genomic DNA PCR screen, the protein extracts of positive cell clones were prepared for protein detection using SDS-PAGE and Western blot (FIG. 1C). The cell clones presented variable levels of Cas9 protein expressi...
example 2
Establishment of HIV Latent Astrocyte Cell Model with Stable Cas9 Expression
[0097]To make a robust HIV latent cell model, a dual fluorescent protein HIV-reporter plasmid-RGH was used. The dual fluorescent protein HIV-reporter plasmid-RGH is described in the Dahabieh et al. (2013) reference, which is incorporated herein by reference in its entirety.
[0098]After RGH pseudovirus infection, when infected cells are in active state, green fluorescence proteins are expressed, and cells are green under fluorescent microscopy (FIG. 2B). When RGH pseudovirus infected cells are in latent stage, only red fluorescent proteins are expressed and cells are red under fluorescent microscopy. According to conventional method, astrocytes were infected with VSVG pseudo-type RGH HIV virus (as described in the Dahabieh et al. (2013) reference) and time-course study was performed of the cell infection status (FIG. 2B). Early during the infection, for example, over first few days, green fluorescent protein e...
example 3
Screening and Testing of gRNAs Targeting HIV Provirus in Latent Astrocyte Cell Lines
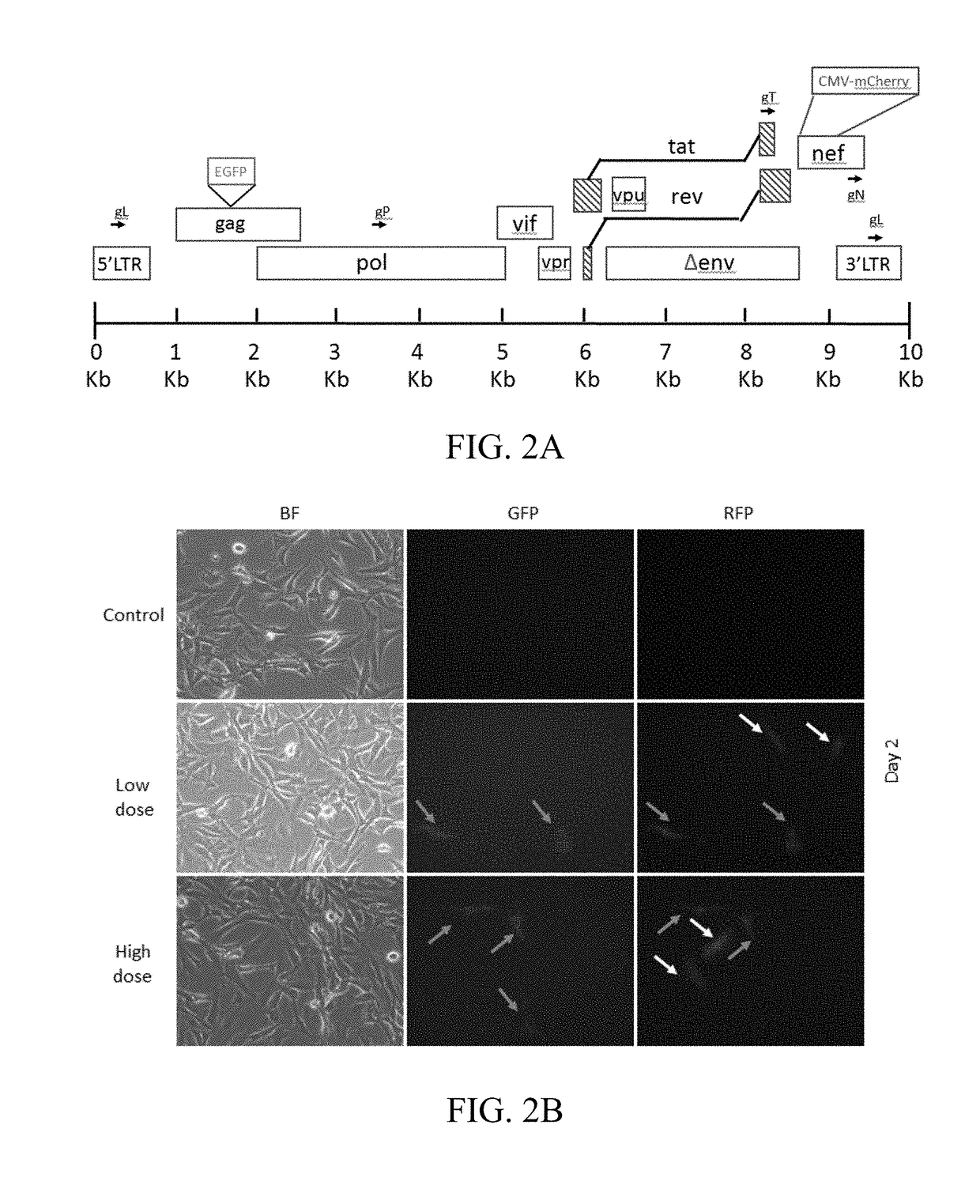
[0099]An in vitro cell platform to screen gRNAs of CRISPR / Cas9 system for the removal of HIV provirus in HIV reporter Cas9 cells is provided. The specificity and gene-editing function of CRISPR / Cas9 system mainly depends on gRNAs. As an initial test, bioinformatics tools were applied to design several gRNAs targeting LTR region, pol gene region, tat gene region and nef gene region as indicated in FIG. 2A.
[0100]RGH Cas9 clones were transfected with those gRNAs either alone or in combination of two (gLTR and gpol; gLTR and gtat; gnef and gpol; gnef and gtat) to delete a DNA fragment which includes the mCherry coding region. Therefore a successful deletion should eliminate the expression of red fluorescent protein (FIG. 2A).
[0101]After transfection, those cells were first imaged with fluorescence microscopy as indicated in FIG. 3A. Reduced expression of red fluorescent protein was observed in gRNAs trea...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pore size | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com