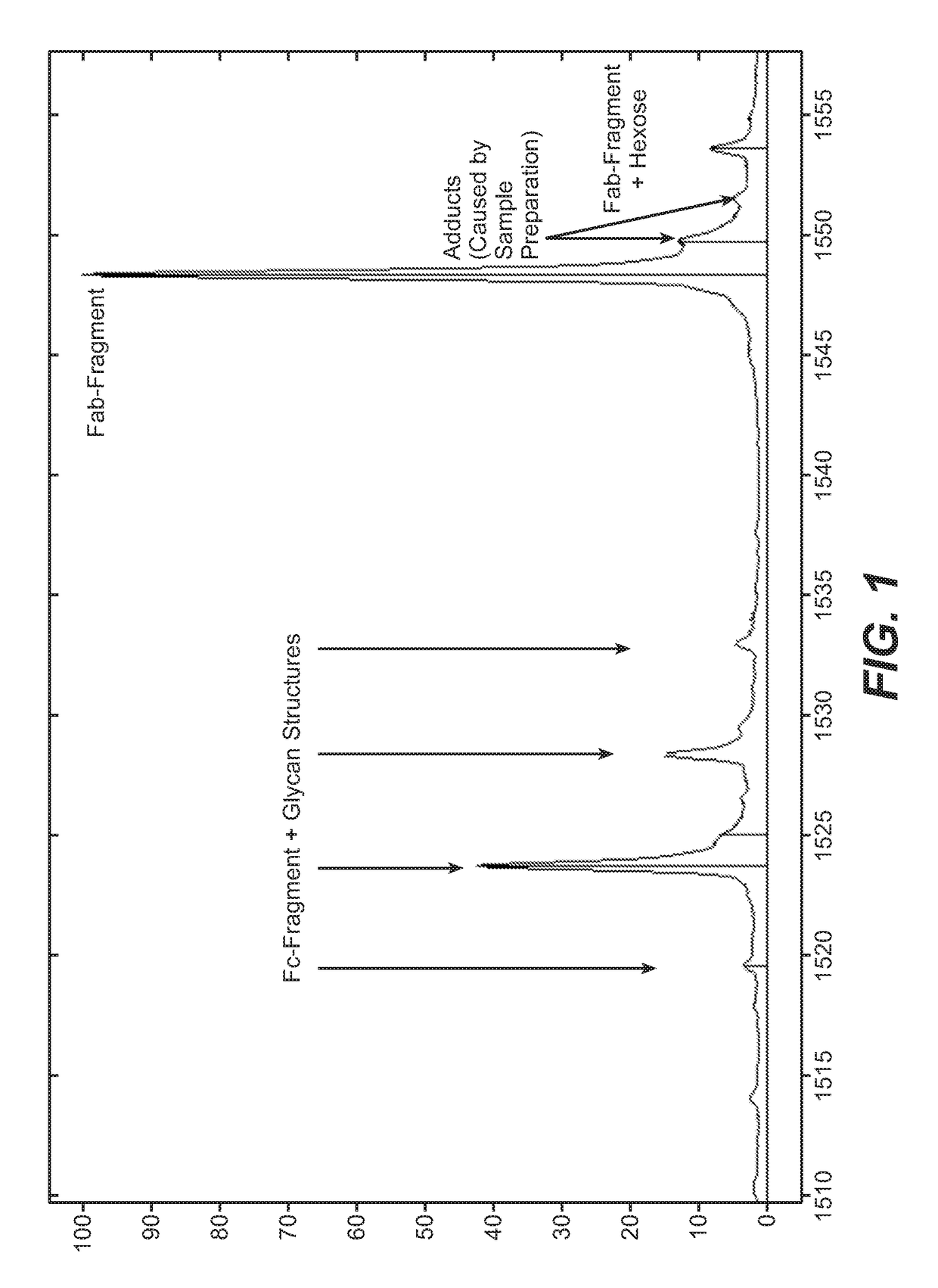
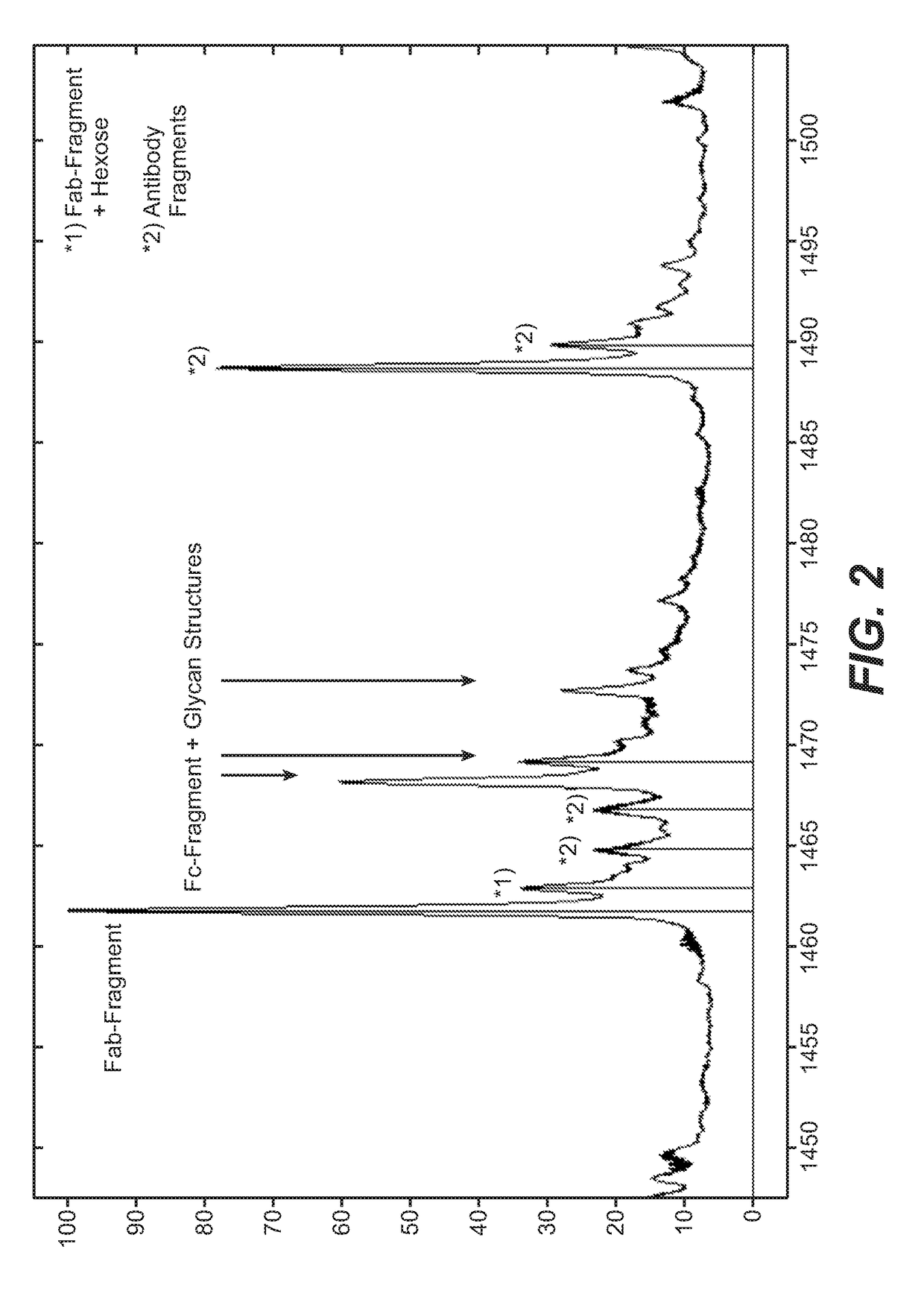
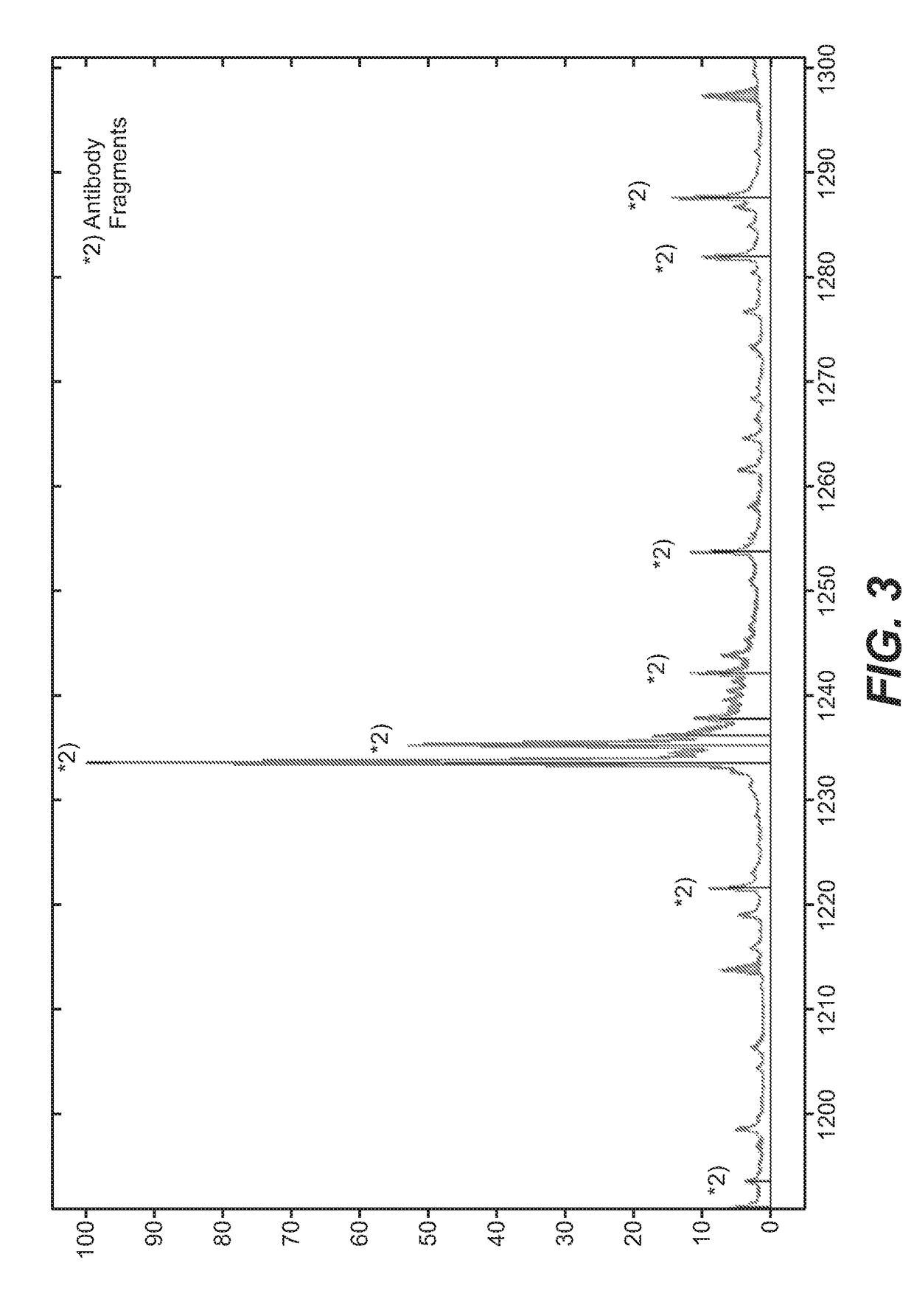
Direct affinity measurement of human igg1 binding multimeric antigens
a multi-mer, affinity-based technology, applied in the field of direct affinity measurement of human igg1 binding multi-mer antigens, can solve the problems of difficult to analyze the diverse chromatographic and electrophoretic bands, separate antibody species, and long time-consuming and laborious problems of applying and optimizing rp-hplc methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Transient Fab Expression and Purification
[0164]The antibody light chain and heavy chain Fd-fragments were ordered as gene syntheses and cloned via unique restriction sites using standard cloning procedures into separate expression vectors for each chain enabling secretory expression in HEK cells growing in suspension. Transfection (1:1 plasmid ratios) into HEK293-F cells (Invitrogen, Cat. No. 510029) was performed according to the cell supplier's instructions using Maxiprep (Qiagen, Cat. No. 12163) preparations of the antibody vectors, Opti-MEM I medium (Invitrogen, Cat. No. 31985) 293fectin (Invitrogen, Cat. No. 31985070), and an initial cell density of 1-2×10E+06 viable cells / mL in serum-free FreeStyle 293 expression medium (Invitrogen, Cat. No. 12338018). Antibody containing cell culture supernatants were harvested after 7 days of cultivation in shake flasks by centrifugation at 14,000× g for 30 min. and filtered through a 0.22 μm sterile filter (Thermo Scientific, Cat. No. 566-0...
example 2
Enzymatic Cleavage of Bevacizumab with Papain
Without Purification:
[0165]The antibody was diluted in 20 mM Histidine, 140 mM NaCl, pH 6.0 to a final concentration of 1 mg / mL, added 2 μL 250 mM L-cysteine (Sigma-Aldrich, Schnelldorf, Germany) and 10.9 μL diluted papain (7.34 U / mL in 20 mM Histidine, 140 mM NaCl, pH 6.0), and incubated 1 h at 37° C.
With Purification:
[0166]The antibody was incubated with Papain (0.8 U / mg mAb; Sigma-Aldrich / Roche) in presence of 5 mM Cystein for 170 minutes at 37° C. To isolate the Fab from non-cleaved antibodies, Fc-fragments and Papain, the mixture was applied to a CaptureSelect IgG-CH1 and MabSelectSuRe affinity chromatography (GE Healthcare) according to manufacturer protocol. Finally, a size exclusion chromatography using a Superdex 75 10 / 300 GL column (GE Healthcare) was performed using 140 mM NaCl, 20 mM histidine (pH 6.0) as running buffer. Protein concentration of the Fab was determined by measuring the optical density (OD) at 280 nm, using the ...
example 3
Enzymatic Cleavage of Bevacizumab with Lysine-Gingipain of Porphyromonas Gingivalis
[0167]GingisKHAN was reconstituted in 200 μL ddH2O resulting in 2000 U / 200 μL, and the 10× reducing agent was freshly prepared in 50 μL ddH2O (final concentration: 20 mM Cysteine) prior to each digestion. 100 μg antibody was diluted to a final concentration of 1 mg / mL in 100 mM Tris, pH 8.0 and subsequently digested with 10 μL GingisKHAN and 11 μL of freshly prepared 10× reducing agent at 37° C. for 1 hour.
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com