Single tube nested type allele specific PCR method
A chain reaction and polymerase technology, applied in the field of molecular biology, can solve the problems of amplification products, high cost, pollution, etc., and achieve the effects of low false positives, simple method and high reliability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
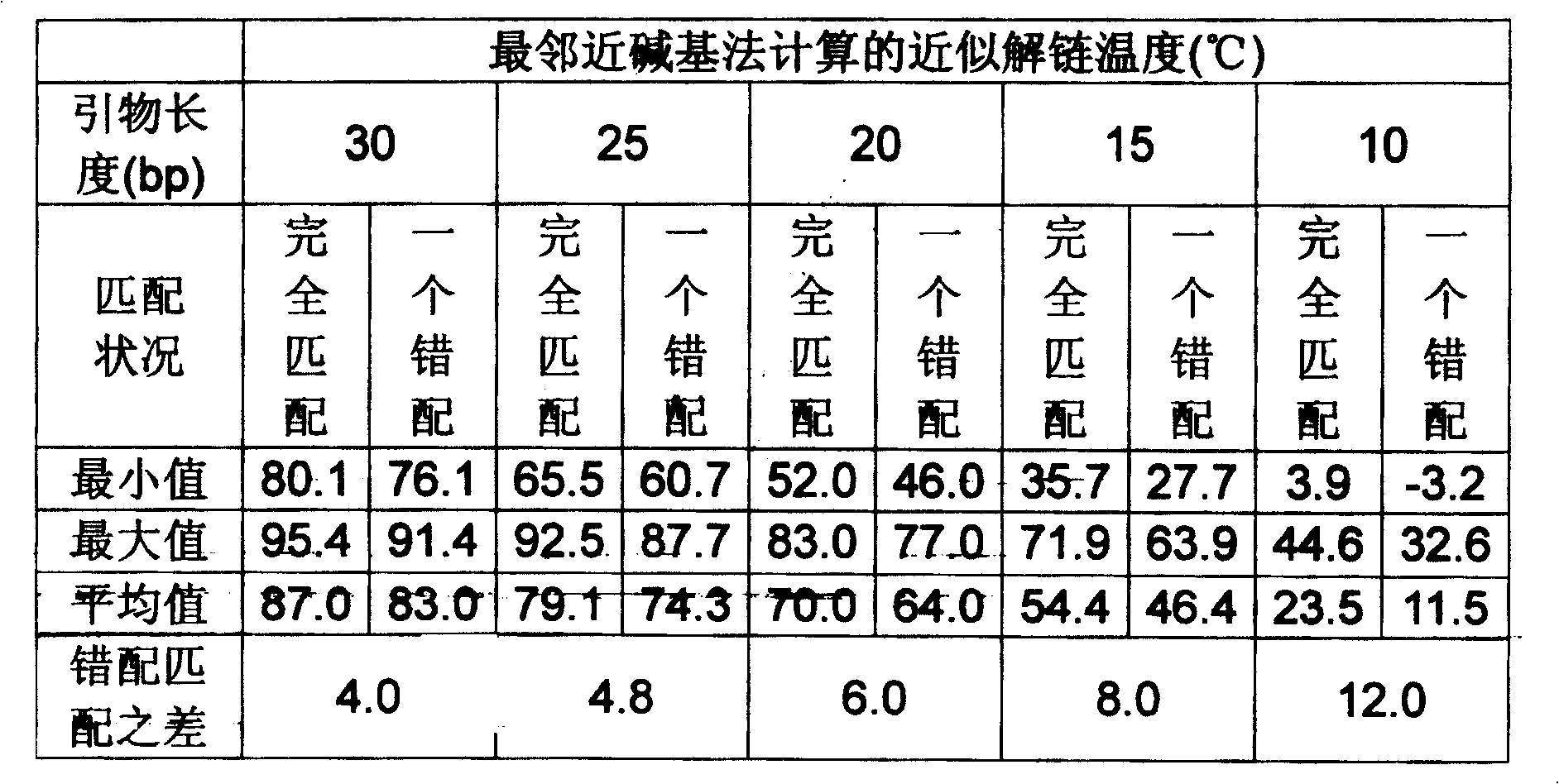
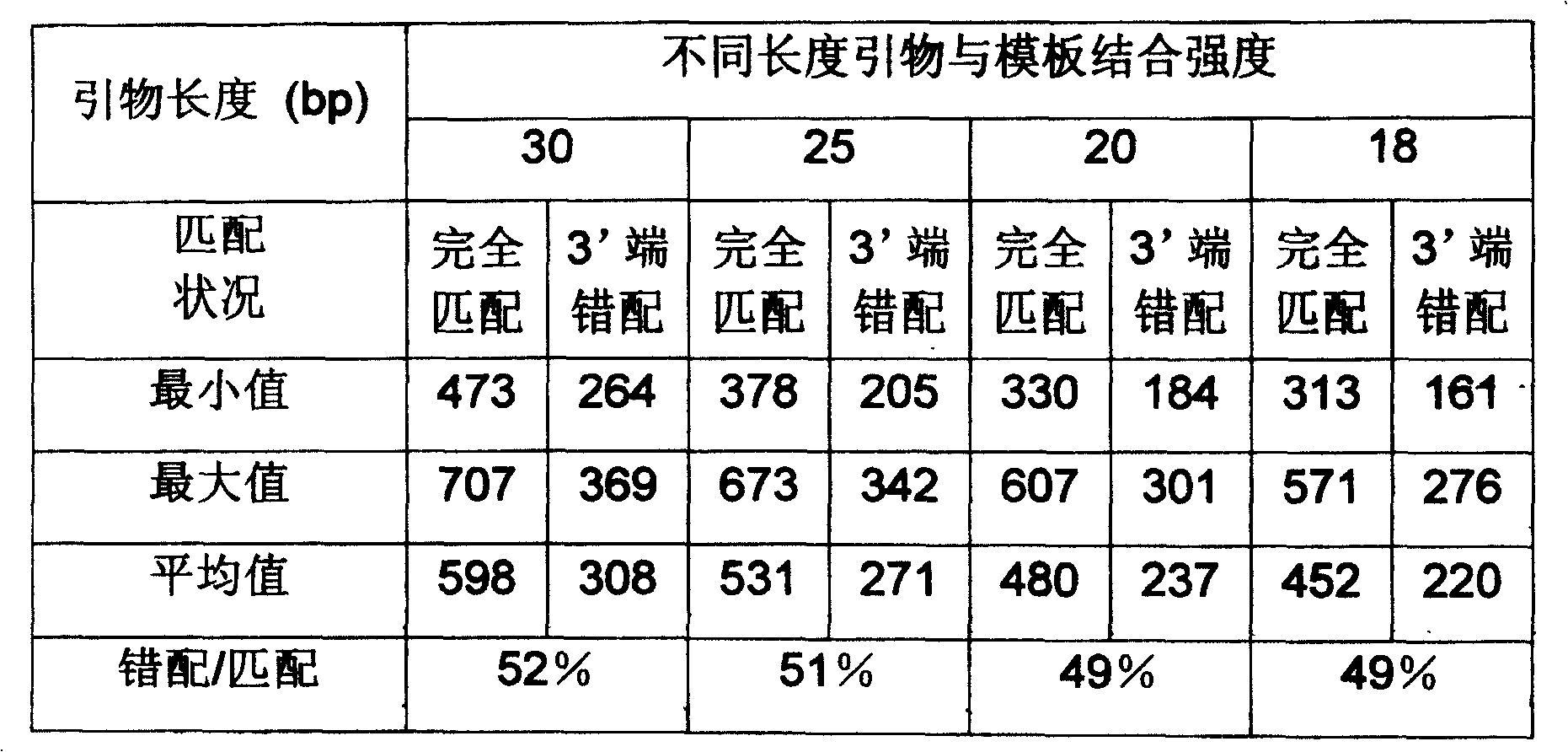
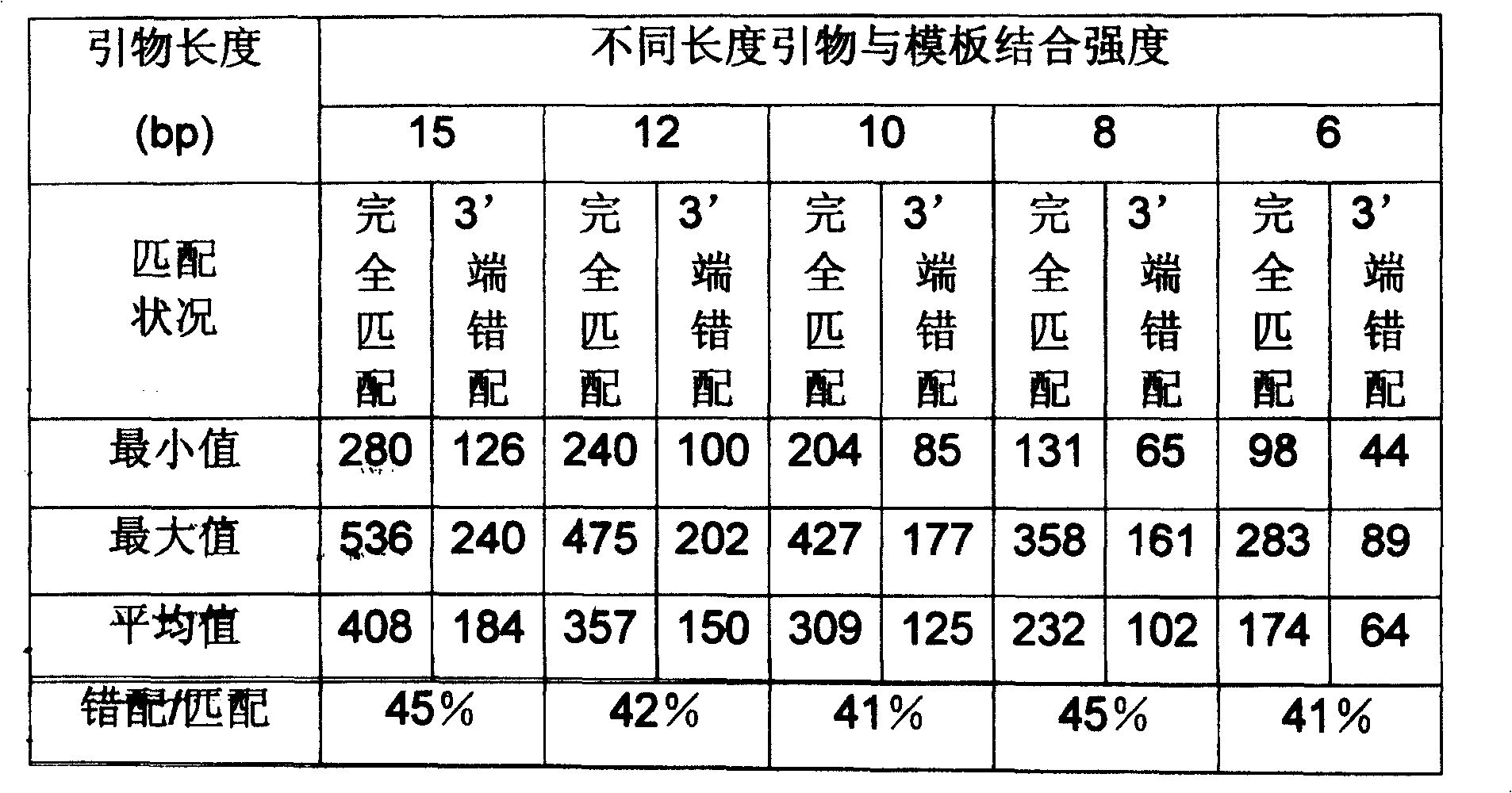
[0043] Example 1 takes the human beta-actomyosin gene (National Center for Bioinformatics GenBank No. BC016045) as the target, assuming that the allele-specific mutation occurs at base 1281 (changed from G to C) or base 1356 (changed from C to G), the primer sequence and properties are shown in Table 6. A1 is the primer for the outer jacket enrichment template, and A2 and A3 are the inner jacket allele-specific PCR primers. The allele-specific primer of A2 is a positive primer, and the third base from the 3' end of the primer is indicated in bold bold letters for the allele-specific mutation; the allele-specific primer of A3 is a reverse primer, and the base at the 3' end of the primer is Allele-specific mutations are indicated in bold boldface.
[0044] Table 6. Example 1 Primer order and characteristics
[0045]
name
Location
long
Spend
degree(℃)
knot with the template
combined st...
Embodiment 2
[0054] Example 2 also targeted the human beta-actomyosin gene (National Center for Bioinformatics GenBank No. BC016045), assuming that the allele-specific mutation occurred at base 1281 (changed from G to C). The primers for coat enrichment template are the same as in Example 1. A4 is the inner set of allele-specific PCR primers. The primer is 20 bases long. The allele-specific primer is a positive primer. The 8th (changed from T to A) and 16th (changed from G to C) counting from the 3' end of the specific primer introduces two mismatched bases, which are underlined. The primer sequence and characteristics are shown in Table 8.
[0055] Table 8. Example 2 primer sequence and characteristics
[0056]
name
Location
length
Spend
(℃)
knot with the template
combined strength
Sequential (5' to 3')
A4 positive
wild
...
Embodiment 3
[0060] Example 3 targets the human glyceraldehyde-3-phosphate dehydrogenase gene (National Center for Biological Information GenBank No. XM_006959). An allele-specific mutation was assumed to occur at base 415 (change from G to C). Primer sequences and properties are shown in Table 9. G1 is the primer for the outer jacket enrichment template, G2 is the inner jacket allele-specific PCR primer, the allele-specific primer is the positive primer, and the fifth base from the 3' end of the primer is indicated in bold bold for the allele-specific mutation .
[0061] Table 9. Example 3 primer sequence and properties
[0062]
name
Location
long
Spend
degree(℃)
knot with the template
combined strength
Sequential (5' to 3')
G1 Positive
343-
366
24
81.4
536
GCT GGC GCT GAG TAC
GTC GTG GAG
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com