Flavonoid-3,5'-hydroxylase gene cloned from moth orchid, its coded sequence and application
A flavonoid, hydroxylase technology, applied in the fields of molecular biology and genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Cloning of Phalaenopsis flavonoid-3’5’-hydroxylase gene
[0037] 1 Phalaenopsis hybrida (Phalaenopsis hybrida) material was collected from the Institute of Vegetable Horticulture, Shanghai Academy of Agricultural Sciences, the variety is Formosa rosa, and the flower colors are purple, white, and yellow. Plant materials are grown under normal conditions in the greenhouse (L / D, 16h / 8h; 25-28°C).
[0038] 2 RNA extraction. Take about 100 mg of fresh material and grind it fully with liquid nitrogen. Add 1mL TriBlue reagent, shake vigorously for 15s, and leave at room temperature for 5min. Add 0.2 mL of chloroform to remove protein, transfer the supernatant to a new centrifuge tube, add an equal volume of isopropanol, mix well, and place at room temperature for 10 minutes. Wash the precipitate with 1 mL of 75% ethanol prepared by DEPC and repeat once. Dry at room temperature for 5-10 minutes, dissolve in 20μL DEPC water, measure OD value, and detect by electrophoresis.
[0039] 3...
Embodiment 2
[0043] Analysis of copy number of Phalaenopsis flavonoid-3’5’-hydroxylase gene
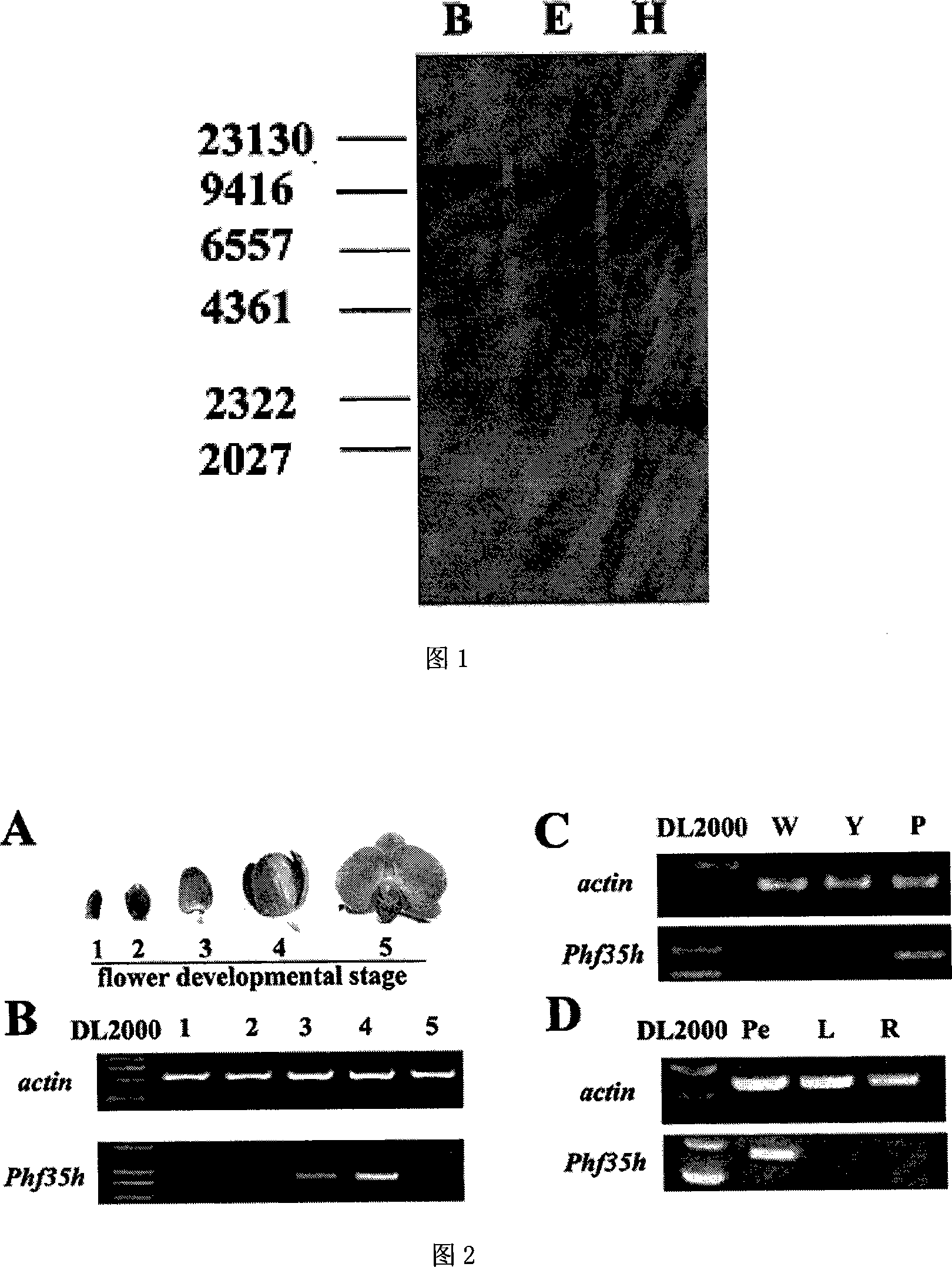
[0044] The Phalaenopsis genome was extracted by CTAB method, and 10 μg DNA was digested with three commercially available enzymes, Bgl II, EcoR V and Hind III, and digested overnight at 37°C. The digested products were separated by electrophoresis using 0.8% agarose gel. Transfer DNA and fix it on nylon membrane. Probe hybridization confirms that it is but a copy. (see picture 1)
Embodiment 3
[0046] Phalaenopsis flavonoid-3'5'-hydroxylase gene eukaryotic expression in petunia plants and phenotype identification of transgenic plants.
[0047] 1 Construction of expression vector. First amplify the complete coding reading frame, and introduce restriction endonuclease recognition sites on the forward and reverse primers (depending on the selected vector) to construct the expression vector. Using the amplified product obtained in Example 1 as a template, after PCR amplification, the Phalaenopsis flavonoid-3'5'-hydroxylase gene was forward cloned into the vector p2301.
[0048] 2 Transfer to Agrobacterium. Prepare Agrobacterium competent, and use heat shock transformation method to introduce plasmid into Agrobacterium.
[0049] 3 Agrobacterium leaf disc method to transform petunia.
[0050] (1) Cut petunia leaves into 0.8×0.8cm (1cm) squares, put them into MSO solution containing Agrobacterium, shaker at 190rpm for 10min.
[0051] (2) Dry the leaves and place them on MS1 c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com