Novel plant strong-salt resistance gene NHXFS1, encoding protein and use thereof
A salt-tolerant gene and plant technology, applied in the field of plant genetic engineering, can solve problems such as unreported, and achieve the effect of improving salt resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Embodiment 1. Arabidopsis, rice, chrysanthemum Na + / H + Cloning of the antiporter gene
[0039] Total RNA was extracted from the young leaves of Arabidopsis thaliana, rice and chrysanthemum with TRizol reagent, and reverse transcription was performed using the total RNA as a template to synthesize the first strand of cDNA. Using the synthesized cDNA as a template and referring to the cDNA sequences of AtNHX1, OsNHX1, and DmNHX1, the following primers were designed (the 5' end contains SmaI and SalI restriction sites respectively):
[0040] AtR(5'-GC GTC GAC TCAAGCCTTACTAAGATCAGGAGG-3')
[0041] AtF(5'-TCC CCCGGG ATGTTGGA TTCTCTAGTG-3')
[0042] OsR(5'-ACGC GTC GAC TCATCTTCCTCCATGGC-3')
[0043] OsF(5'-TCC CCCGGG ATGGGGATGGAGGTGGCG-3')
[0044] DmR (5'-GC GTC GAC TTAGTTTCTTTCTTTCATCTTC-3')
[0045] DmF(5'-TC CCCCGG GATGGTGTTCGATTC-3')
[0046] Arabidopsis Na + / H + Antiporter gene AtNHX1, rice Na + / H + Antiporter genes OsNHX1 and chrysanthemum Na...
Embodiment 2
[0047] Example 2. DNA shuffling of AtNHX1, OsNHX1, DmNHX1 genes
[0048] 1) Preparation of starting materials Using pGM-T-AtNHX1, pGM-T-OsNHX1, and pGM-T-DmNHX1 as templates, use PfuDNA polymerase to PCR amplify the three genes, and use them as starting materials for DNA shuffling after purification .
[0049] 2) Random enzyme digestion with DNase I Determine the concentration of purified DNA by ultraviolet absorption method, mix AtNHX1, OsNHX1, and DmNHX1 with a content of 9ug, so that the ratio of the three gene substances is 1:1:1. Take 40ul of mixed AtNHX1, OsNHX1, DmNHX1 and add it to 50μl enzyme digestion reaction system (10mM Tris-HCl, pH7.5, 50mM MnCl 2 ), react at 15°C for 10 min. Add DNase I 0.15U, mix well, 15°C, 1min16s, 90°C, 10min. The product is electrophoresed through 2.5% agarose gel, and the gel containing 100-200bp fragments is excised and recovered.
[0050] 3) PCR without primers Take 1 μg of recovered small fragments and add to 50 μl reaction system (1...
Embodiment 3
[0053] Example 3. Construction of expression shuffling library and resistance to high salt Na + / H + Screening of antiporter gene NHXFS1
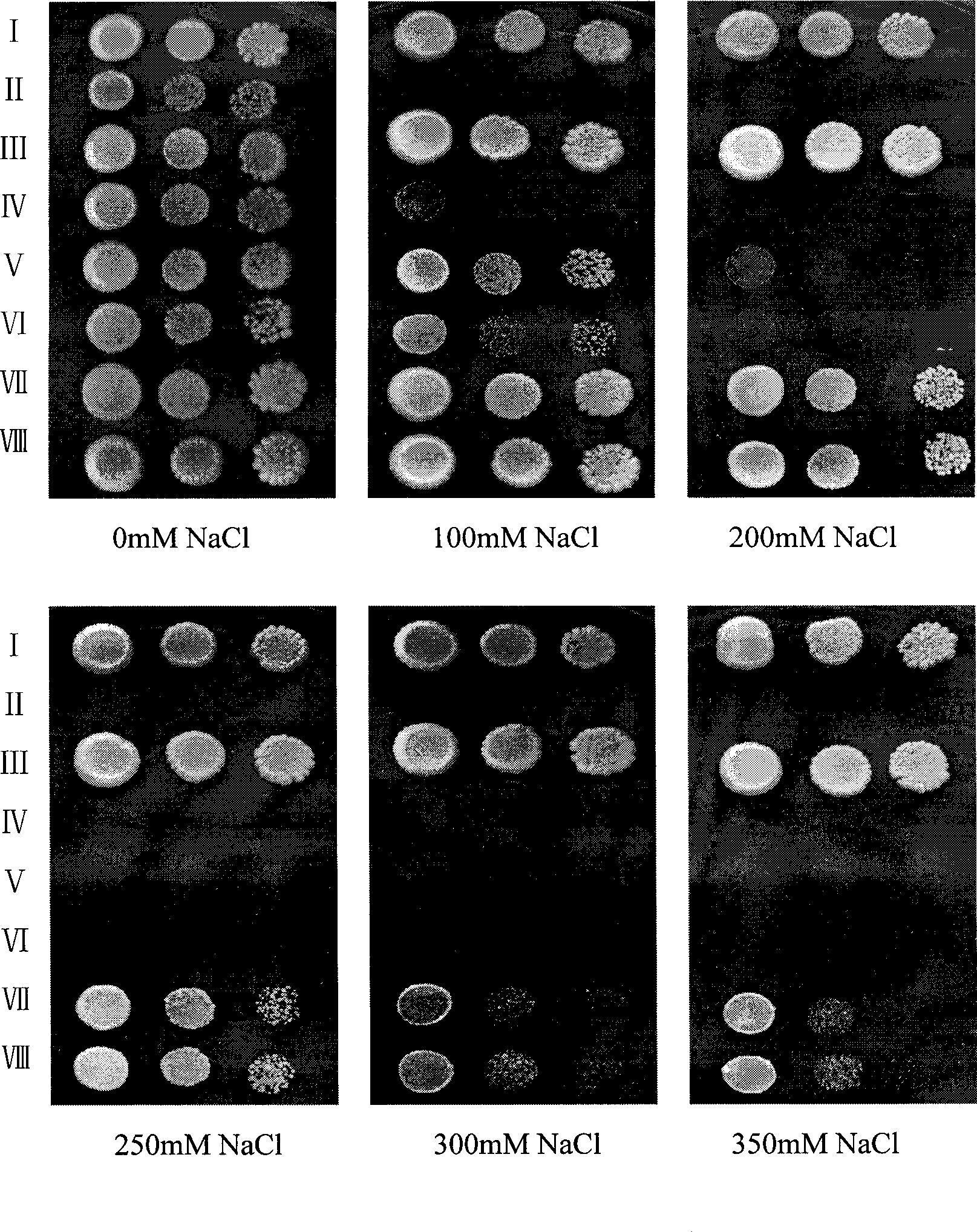
[0054] The recombinant product obtained in Example 2 was digested with Sal I and Sma I, purified and connected to the same digested yeast expression vector pYPGE15, and the constructed expression shuffling library was introduced into the yeast mutant strain W303-1B△ by the lithium acetate method ena1-4::HIS3 △nhx1::TRP1, spread on the APG selection medium without uracil, culture at 30°C for 2 days, and select Na + / H + The yeast strain of the antiporter gene was further screened by increasing the NaCl concentration, and finally a normal growth yeast mutant was obtained on the selection medium with a concentration of 200mM NaCl, which contained a new plant Strong salt tolerance gene NHXFS1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com