Detection of bacterium by utilizing dnaj gene and use thereof
A technology of bacteria and genes, applied in the determination/inspection of microorganisms, recombinant DNA technology, resistance to vector-borne diseases, etc., can solve the problem of large sequence differences (Non-patent document 8, high polymorphism level at the species level).
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
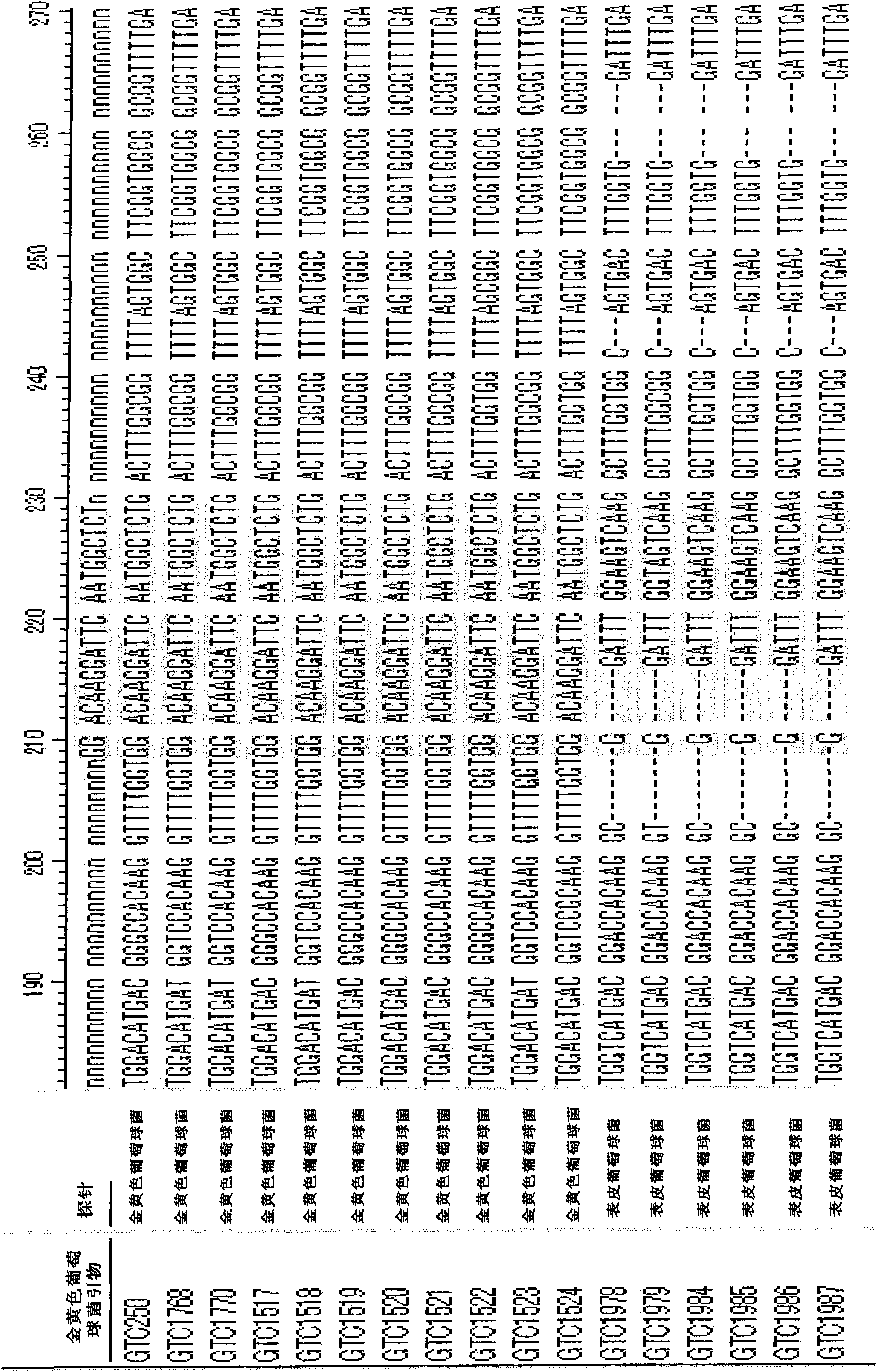
[0362] Staphylococcus aureus (S.aureus) of the genus Staphylococcus is an important species that causes food poisoning, and it is known that in terms of 16S rDNA, it has a similarity of 99.5 to the closely related bacteria Staphylococcus epidermidis (S. %( Figure 1A ), so it is impossible to design strain-specific primers, but for the DnaJ gene, the similarity of these two strains is only 81.1% ( Figure 1B ). Therefore, primers can be easily designed for the DnaJ gene. Moreover, the primer sequence is relatively conservative among the 11 strains of Staphylococcus aureus (S.aureus) whose sequence has been determined ( figure 2 ). In this example, based on the determined DnaJ sequences of 9 pathogenic bacteria causing food poisoning, primers for these pathogens were prepared. The primer sequences for each pathogen are as follows.
[0363] Primers for Staphylococcus aureus
[0364] SEQ ID NO: 13:5-GGACAAGGATTCAATGGCTCT
[0365] SEQ ID NO: 14:5-TTGCGGTGCATTTGGATCTCT
[0...
Embodiment 2
[0410] Instead of DNA of Staphylococcus aureus (S. aureus) used in Example 1, DNA of the following strains was used, and a mixed primer set was used under the same conditions. DNA amplified with a Light Cycler (Roche) capillary was monitored by an increase in CYBR green fluorescence intensity. The results are shown in Figure 4 .
[0411] [Strains used]
[0412] 1. Campylobacter jejuni GTC 0259 strain
[0413] 2. Vibrio cholerae (Vibrio cholerae) GTC 0037 01 E1 Tol type
[0414] 3. Vibrio parahaemolyticus GTC 0056 (=CDC A3308)
[0415] 4. Escherichia coli (Escherichia coli) GTC 1061 0157 (VT1 and VT2 positive)
[0416] 5. Listeria monocytogenes (Listeria monocytogenes) GTC0149 (=ATCC15313) type strain
[0417] 6. Salmonella enterica var. Typhimurium GlFU 11566 ST-1
[0418] 7. Yersinia enterocolitica (Yersinia enterocolitica) GTC 0127 strain
[0419] 8. Clostridium perfringens GTC 0785 (=ATCC13124)
[0420] Such as Figure 4 As shown, it was confirmed that the DNA of...
Embodiment 3
[0422] (Amplification of the DnaJ gene of Vibrio bacteria)
[0423] A primer solution obtained by mixing a universal forward primer (SEQ ID NO: 43) for bacteria belonging to the genus Vibrio (SEQ ID NO: 43) and a species-specific reverse primer within the same genus other than those described below was prepared in the following manner.
[0424] Vibrio universal primer (SEQ ID NO: 43) 2 μM
[0425] Vibrio cholerae (SEQ ID NO: 44) 1 μM
[0426] Vibrio mimicus (SEQ ID NO: 47) 1 μM
[0427] Vibrio vulnificus (SEQ ID NO: 53) 1 μM
[0428] Vibrio fluvialis (SEQ ID NO: 134) 1 μM
[0429] Vibrio parahaemolyticus (SEQ ID NO: 50) 1 μM
[0430] Vibrio alginolyticus (SEQ ID NO: 58) 1 μM
[0431] In addition, this primer solution was used to prepare a PCR reaction solution in the following manner.
[0432] Primer mixed solution (forward μM, reverse each 1μM): 2μl
[0433] 2×CYBER premixed Hotstart Ex Taq (Takara): 10μl
[0434] DNA of each Vibrio (single): 5ng
[0435] Water was a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com