Phosphatidate phosphatase homolog and use of the same
A phosphatidic acid phosphatase, fatty acid technology, applied in application, hydrolase, food science and other directions, can solve problems such as no function
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
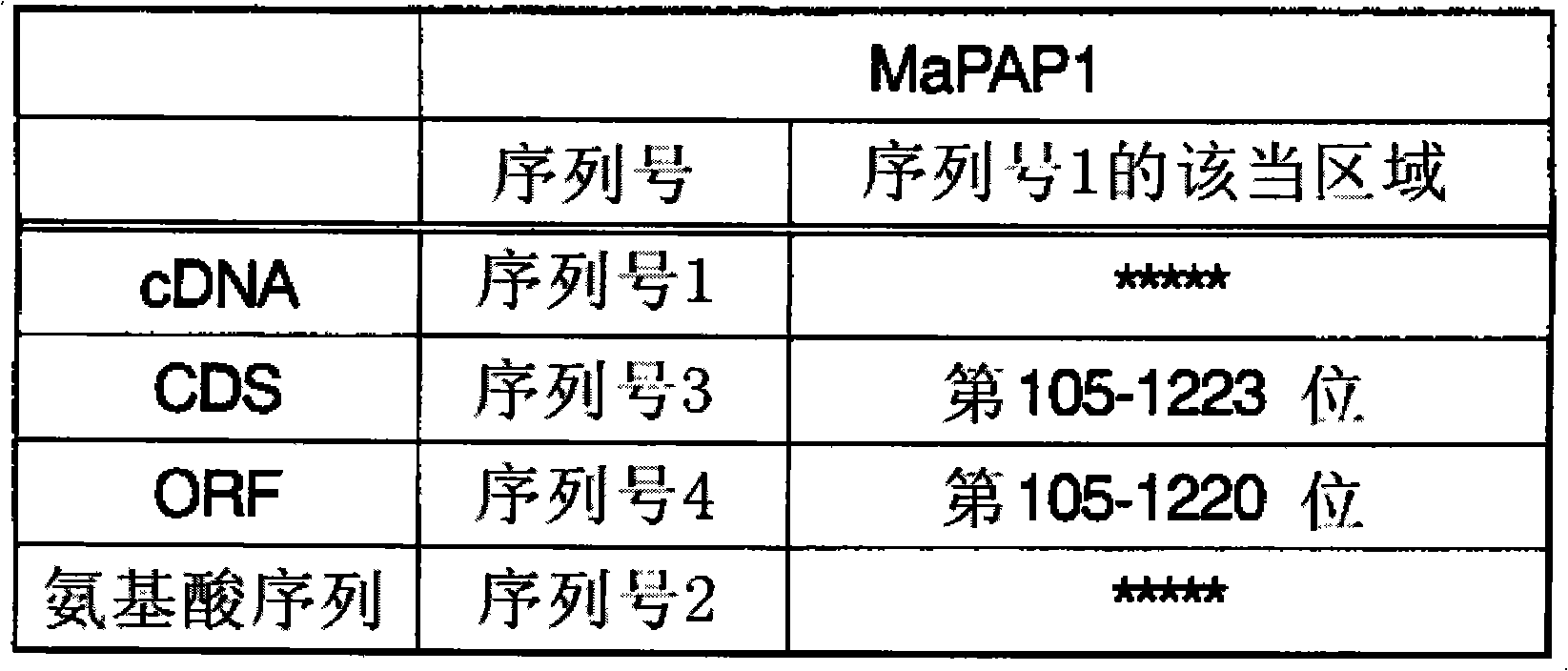
[0249] PAP expression vector construction and transformant preparation
[0250] The present invention also provides a recombinant vector containing the nucleic acid encoding the PAP of the present invention. The present invention further provides a transformant transformed with the above-mentioned recombinant vector.
[0251] Such recombinant vectors and transformants can be obtained as follows. That is, the plasmid having the nucleic acid encoding the PAP of the present invention is digested with restriction endonucleases. The restriction endonucleases used are, for example, EcoRI, KpnI, BamHI, SalI, etc., but are not limited thereto. And the ends can also be blunted by T4 polymerase treatment. Purify the base sequence fragment after enzyme digestion by agarose gel electrophoresis. By integrating this nucleotide sequence fragment into an expression vector by a known method, a PAP expression vector can be obtained. The expression vector is introduced into the host to pr...
Embodiment 1
[0303] (1) EST analysis
[0304] Mortierella alpina (M.alpina) 1S-4 strain was inoculated into 100 ml of medium (1.8% glucose, 1% yeast extract, pH 6.0), and pre-cultivated at 28° C. for 3 days. Add 5L medium [1.8% glucose, 1% soybean powder, 0.1% olive oil, 0.01% Adecanol, 0.3% KH to 10L culture tank (Able Co., Tokyo) 2 PO 4 , 0.1%Na 2 SO 4 , 0.05% CaCl 2 2H 2 O, 0.05% MgCl 2 ·6H 2 0, pH 6.0], inoculate all the precultures, and culture with aeration and agitation under the conditions of 300rpm, 1vvm, and 26°C for 8 days. Glucose corresponding to 2%, 2%, and 1.5% was added on the 1st, 2nd, and 3rd day of culture, respectively. The bacterial cells were recovered at each stage of the 1st, 2nd, 3rd, 6th, and 8th days of culture, and the total RNA was prepared by the guanidine hydrochloride / cesium chloride method. Use Oligotex-dT30 mRNA Purification Kit (TaKaRa Bio), to purify poly(A) from total RNA + RNA. Using the ZAP-cDNA Synthesis Kit (STRATAGENE), the cDNA libr...
Embodiment 2
[0308] (1) Cloning of PAP homologous gene (MaPAP1)
[0309] In order to obtain a cDNA fragment containing the full-length ORF of the PAP homologous gene, primers were designed as follows:
[0310] Primer D-1: CATGGGTTGCTTCGCGCGCAAGACG (SEQ ID NO: 5)
[0311] Primer D-2: CGAAGCCGGCAAAGGCGGCAGTC (SEQ ID NO: 6)
[0312] A PCR reaction was carried out using ExTaq (TaKaRa Bio) using the cDNA library on day 8 as a template and using the above-mentioned primers. The obtained 0.64 kbp DNA fragment was subjected to TA cloning using TOPO-TA cloning Kit (INVITROGEN), and the base sequence of the insert was determined.
[0313] As a result, it was confirmed that a DNA fragment including the 113th to 744th base sequence of SEQ ID NO: 1 was cloned, and this plasmid was designated as pCR-2051-P. Then, PCR reactions were carried out using the plasmid pCR-2051-P as a template and using the same primers as above. ExTaq (TaKaRa Bio) was used in the reaction, but the PCR labeli...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com