Construction of lactobacillus acidophilus S-layer protein surface display system
A surface display system, Lactobacillus acidophilus technology, applied in biochemical equipment and methods, material inspection products, applications, etc., can solve the problem of no protein expression and achieve the effect of weakening the degradation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
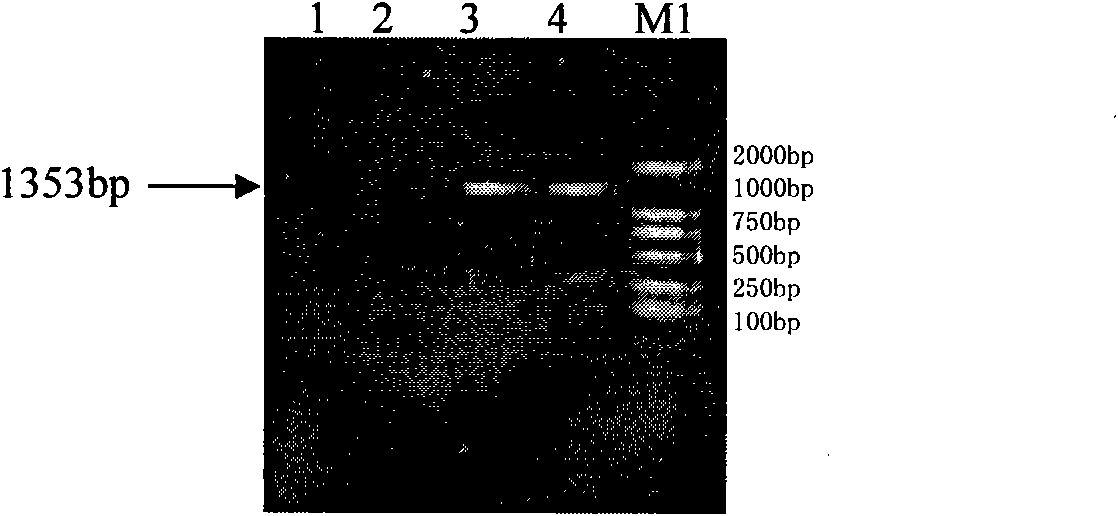
[0078] (1) Acquisition of the S-layer SlpA gene of Lactobacillus acidophilus
[0079] 1 Materials and reagents
[0080] (1) strain
[0081] Lactobacillus acidophilus strain (Lactobacillus acidophilus 1.1878), Escherichia coli (E. coli JM109).
[0082] (2) Enzymes and main biochemical reagents
[0083] pMD18-T Vector System, Restriction Enzyme Sac I, Kpn I, dNTP, Ex Taq DNApolymerase, RNaseA, λ-Hind III Digest Marker, DL2000 Marker, Lysozyme, DNA Gel Recovery and Purification Kit, IPTG, Agar sugar.
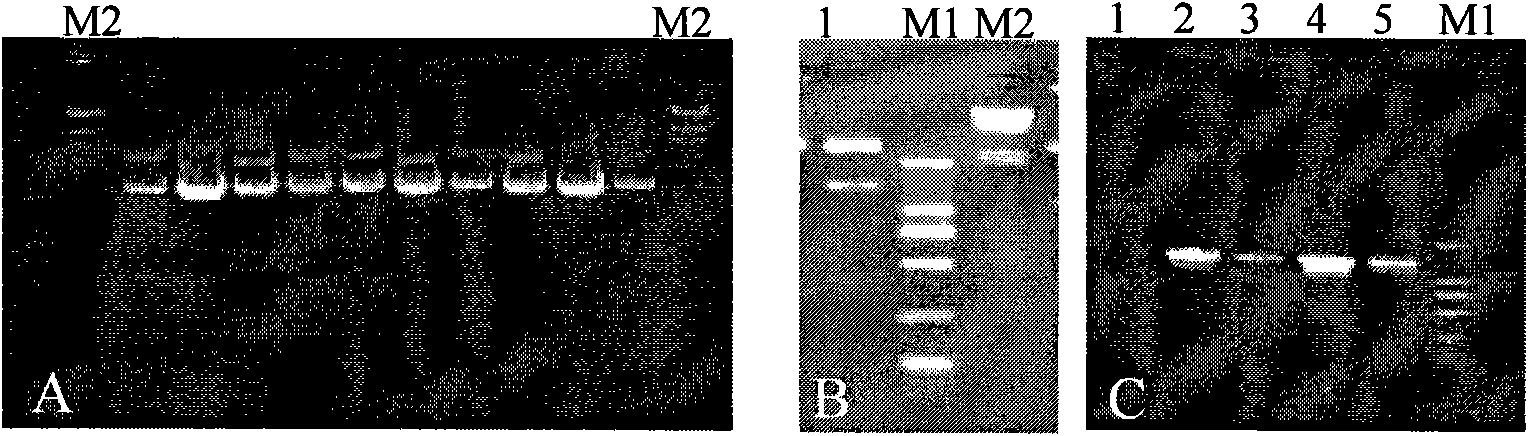
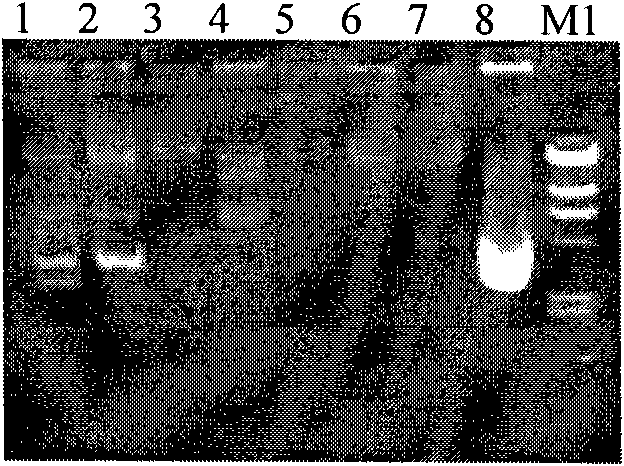
[0084] 2 PCR amplification of SlpA gene
[0085] After the revived Lactobacillus acidophilus strains were cultured overnight at 37°C constant temperature anaerobic culture, a single colony was picked and inoculated in fresh MRS liquid medium for cultivation. 600 When the value is 0.6-0.8, collect the bacteria, use the chromosome extraction kit, and extract the chromosome DNA according to the instructions. According to the sequence of the published S-layer gene, a pair of prim...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com