SiRNA for specifically inhibiting ECHS1 gene expression and application thereof
A transgenic mouse and purpose technology, applied in the field of siRNA, can solve the problems of slow modeling speed, high price, poor model stability, etc., and achieve the effect of increasing lipid droplets and promoting growth
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Embodiment 1, siRNA design and synthesis
[0031] 1. Synthesis of ECHS1 siRNA
[0032]Obtain the full-length sequence of mouse ECHS1 mRNA (NM_053119.2; sequence 3 in the sequence table) from the database of the National Center for Biotechnology Information (NCBI), and design and synthesize it based on the principle of RNAi and design software. And an effective 21nt siRNA against the mouse ECSH1 gene was screened out. A BLAST comparison check was performed to ensure that there was no homology to other genes.
[0033] The sequence of ECHS1 siRNA is: 5'-GCCUUUGAGAUGACGUUAAtt-3' (sense strand) (sequence 1);
[0034] 3'-gtCGGAAACUCUACUGCAAUU-5' (antisense strand) (SEQ ID NO: 2).
[0035] Chemical synthesis was carried out by Shanghai Gemma Pharmaceutical Technology Co., Ltd. (factory address: 602, No. 1011 Halei Road, Zhangjiang, Pudong, Zip Code: 201203): using NTP as raw material, using ABI3900 nucleic acid synthesizer to chemically synthesize single-stranded RNA, and f...
Embodiment 2
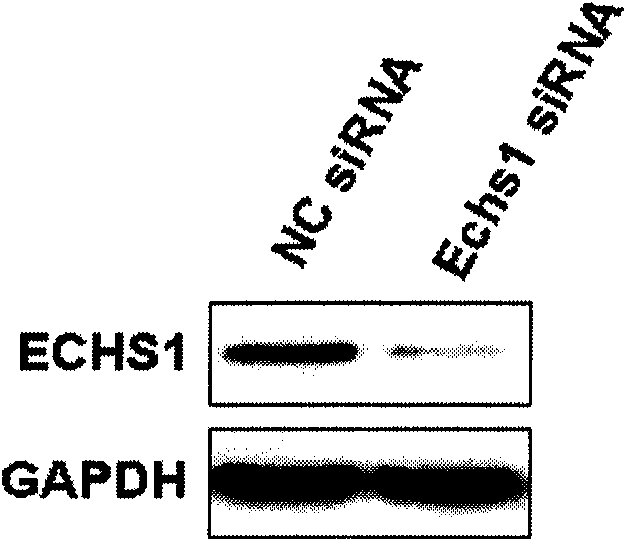
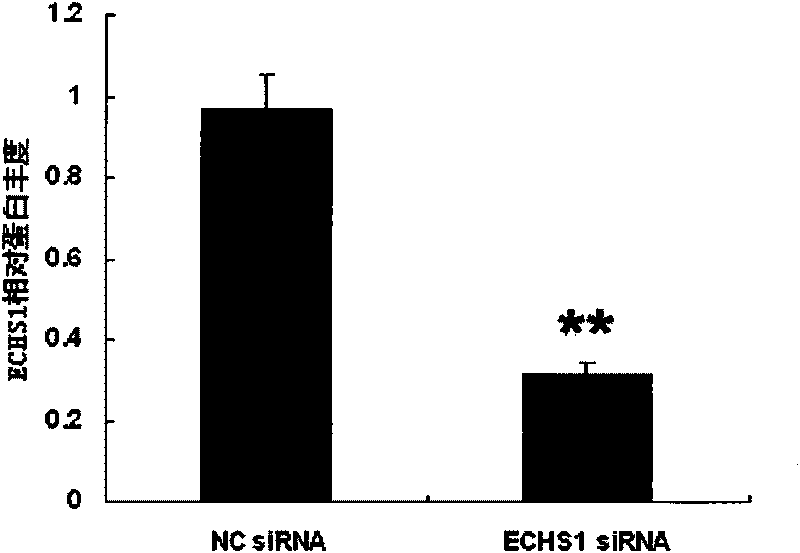
[0040] Example 2, Effect of ECHS1 siRNA on the expression of ECHS1 gene in AML-12 cells in vitro
[0041] 1. Group transfection
[0042] Spread AML-12 cells evenly in a 24-well plate and divide them into 4 groups:
[0043] High-fat experimental group: containing 10% (volume percentage) inactivated newborn bovine serum, 100U / ml penicillin and 100mg / ml streptomycin, sodium oleate of 666 μ mol / L and sodium palmitate of 333 μ mol / L DMEM / F12 medium; transfect 200 pmol of ECHS1 siRNA of Example 1.
[0044] High-fat control group: containing 10% (volume percentage) inactivated newborn bovine serum, 100U / ml penicillin and 100mg / ml streptomycin, sodium oleate of 666 μ mol / L and the sodium palmitate of 333 μ mol / L DMEM / F12 medium; 200 pmol negative control siRNA of Example 1 was transfected.
[0045] Normal experimental group: DMEM / F12 medium containing 10% (volume percent) inactivated neonatal bovine serum, 100 U / ml penicillin and 100 mg / ml streptomycin; transfected with 200 pmol of...
Embodiment 3
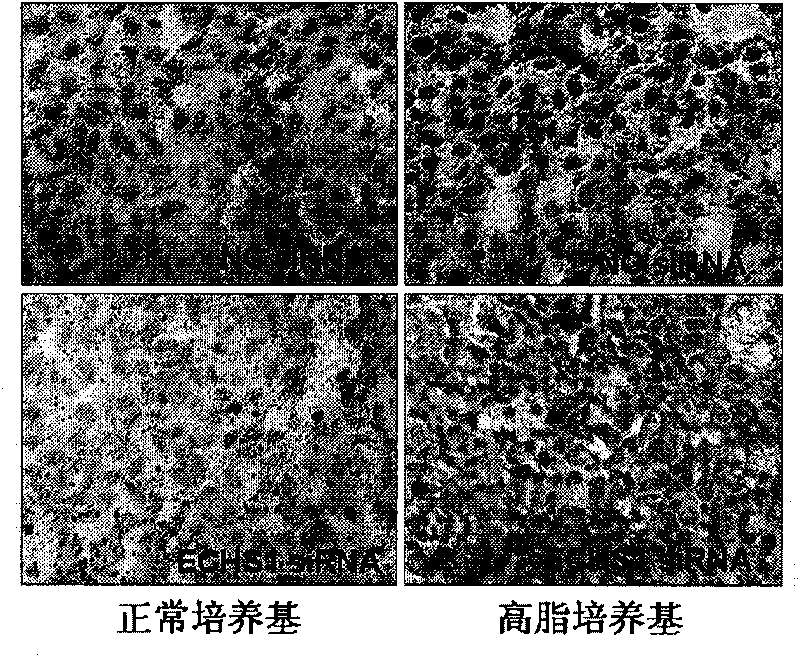
[0051] Example 3, Effect of ECHS1 siRNA on AML-12 Cell Steatosis in Vitro
[0052] 1. Group transfection
[0053] AML-12 cells were spread evenly in 12-well plates and divided into 4 groups. Concrete grouping is with the step one of embodiment 2.
[0054] 2. The effect of ECHS1 siRNA on lipid change of AML-12 cells in vitro
[0055] The cells after step 1 were subjected to the following experiments 1 or 2 or 3, respectively.
[0056] 1. Oil red dyeing
[0057] Add 4% paraformaldehyde to a 12-well plate and fix at 4°C for 30 min, wash twice with distilled water and then add oil red O dilution (oil red O 0.5g dissolved in isopropanol 100ml saturated solution and distilled water by 3: 2 diluted, left to stand for 5-10min and filtered for use), dark stained for 10-15min, washed once with 60% ethanol and differentiated under the microscope until the interstitium was clear, washed twice with Mayer’s hematoxylin counterstained nuclei for 8 seconds, washed twice Afterwards, inver...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com