High-efficiency ammonium-excreting combined azotobacter strain
A high-efficiency technology for nitrogen fixation, applied in the direction of bacteria, microorganism-based methods, microorganisms, etc., can solve the problems of limiting the development and application of nitrogen-fixing bacteria
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1a
[0037] Example 1 amtB mutant strain construction
[0038] 1) Amplification of amtB1 gene homologous fragment
[0039] Using A1501 total DNA as a template, respectively
[0040] 5′-ACAGGATCCTCTGTTAAGCCCAGGCTT-3′
[0041] 5′-GGGAGGCTTATTGAAGTTGGTGACGCC-3
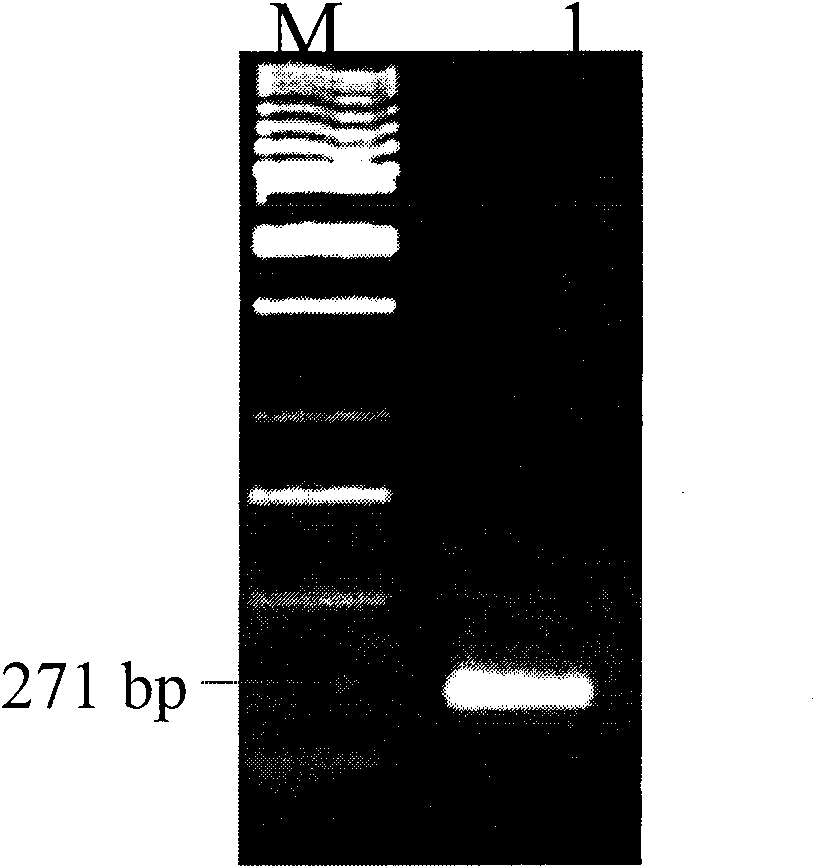
[0042] Carry out PCR amplification for primers, amplify the DNA fragment that is used for constructing amtB gene mutant strain, the product is verified by agarose gel electrophoresis ( figure 1 ).
[0043] 2) Construction of amtB1 gene integration mutants
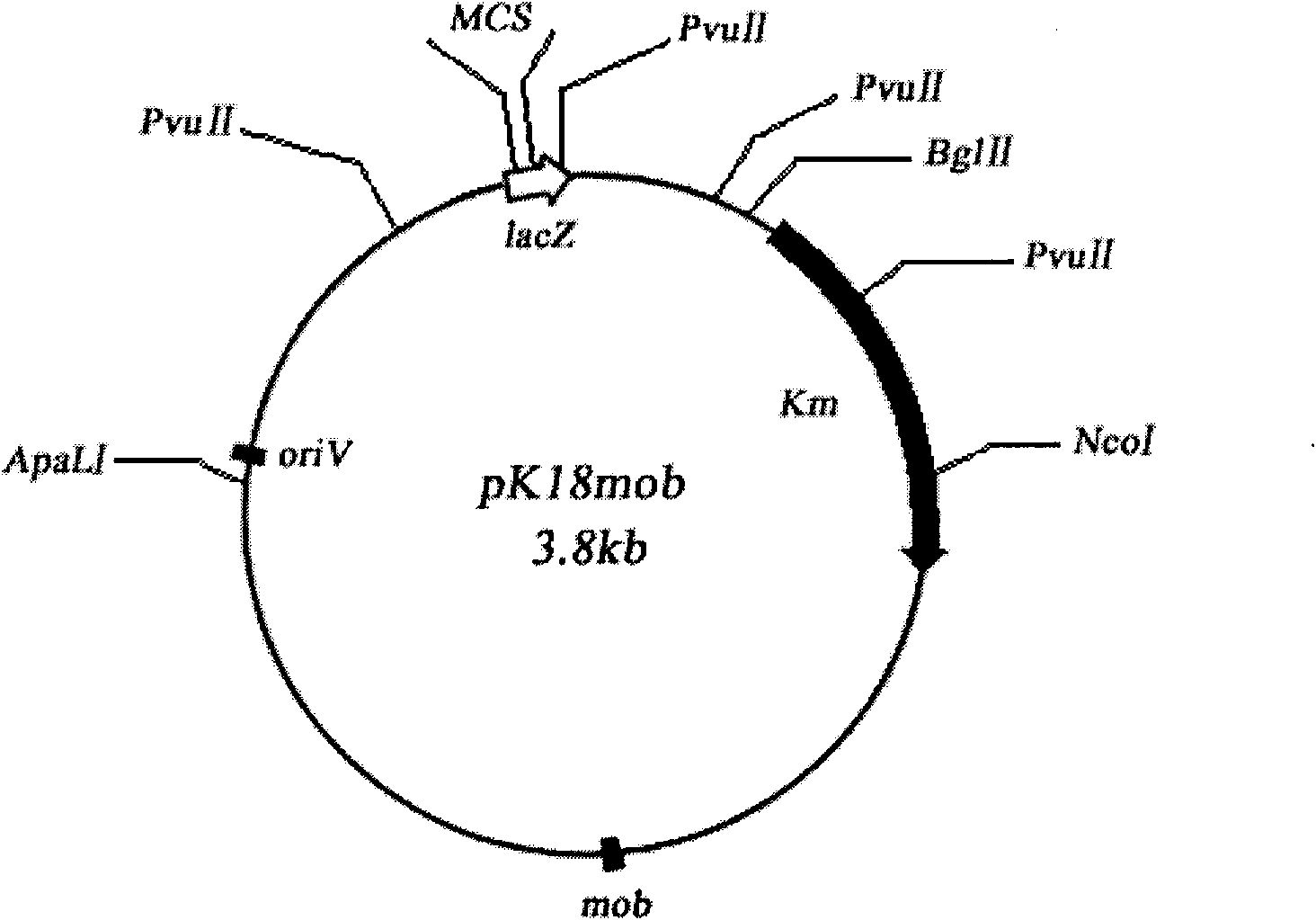
[0044] Homologous fragments obtained by PCR were digested and connected to the multiple cloning sites of the digested plasmid pK18mob to obtain recombinant plasmids. After the recombinant plasmid was identified by enzyme digestion, it was introduced into the recipient strain P. stuteri A1501 through triparental conjugation, so that the plasmid had homologous single exchange on the target gene, integrated into the chromosome, and the triparental zygote was obtained by ...
Embodiment 2
[0049] Example 2 Construction and verification of 1561 / pVA3 strain
[0050] 1. Cloning of nifA gene molecules
[0051] The genome of Pseudomonas stutzeri A1501 was extracted. Using the extracted genome as a template, PCR amplification was performed. The primers used were synthesized according to the 5' and 3' end sequences of the nifA gene, and the sequences of the two primers were:
[0052] 5' CCGAAGCTTTCAGATCTTGCGCATATGA 3' and
[0053] 5' CCGAAGCTTACGGTGCATATCGATAGC 3',
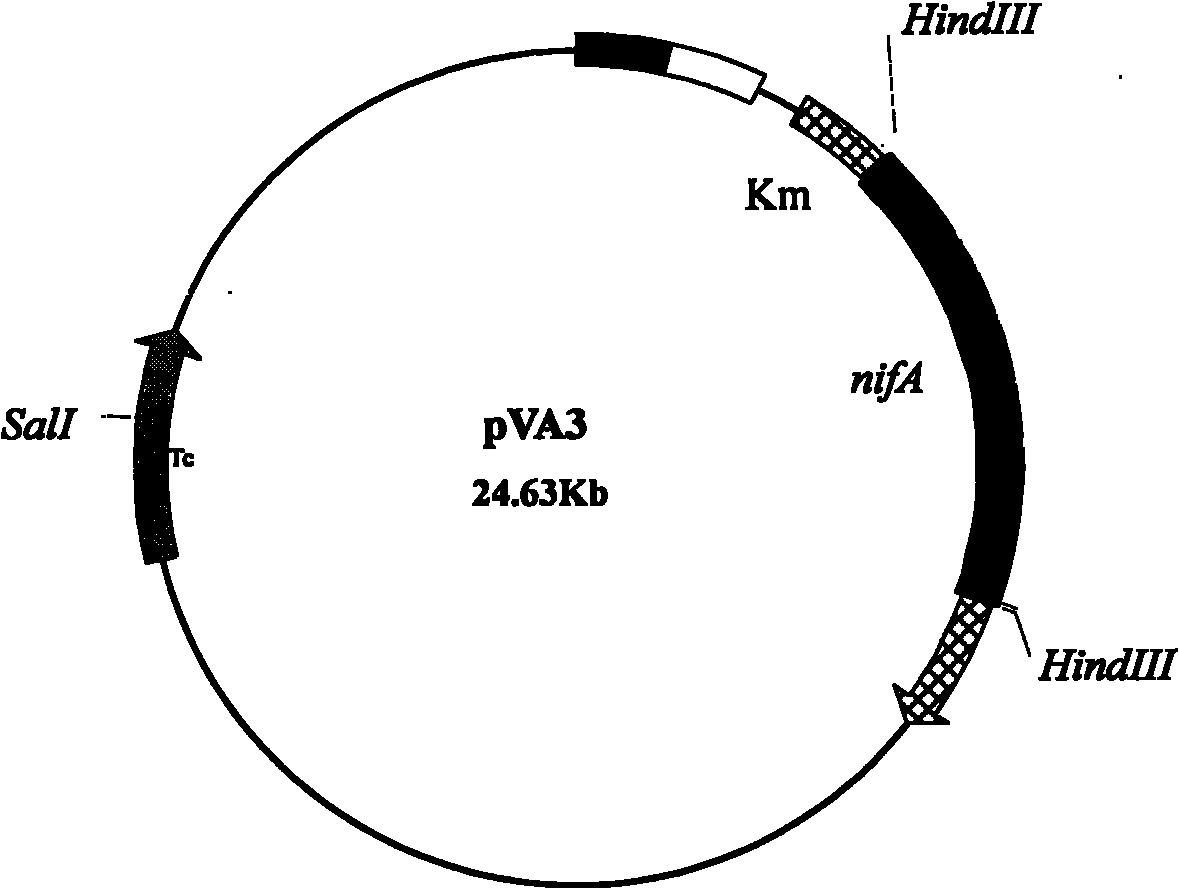
[0054] The about 1124bp fragment of phytase phyc gene was recovered by electrophoresis. The complete nifA gene is obtained by the PCR method, and includes a nucleotide sequence about 70 bp upstream of the coding region of the nifA gene, and a target fragment of 1.64 kb in length. Digest with HindIII and recover the target fragment, and connect it with the shuttle vector pVK100 digested with the same enzyme. Tc s Km r Resistance screening, plasmid extraction, HindIII and EcoRI digestion identificati...
Embodiment 3
[0058] The ammonium secretion comparison of embodiment 3 engineering bacterium 1561 / pVA3 and wild-type bacterial strain
[0059] 1) Determination of ammonium secretion by strains
[0060] NH 4 + The determination of concentration is according to indophenol blue colorimetric method (Bender et al.1977): A solution (100ml): 5g phenol; 0.025g Na-nitroprusside (Na 2 Fe[CN] 5 NO2H 2 O); B solution (100ml): 62.5ml 1M NaOH; 3.3ml NaOCl (available chlorine 5.25%). Take 100 μl bacterial culture supernatant, add 100 μl A solution, then add 100 μl B solution, observe the color depth of the reaction solution after 20 minutes to judge whether there is NH 4 + . Appropriately expand the volume and draw NH 4 + Standard curve (serial dilution of 1M NH 4 + 0.1, 0.2, 0.4, 0.6, 0.8, 1.0, 1.5, 2.0mM; after adding A and B solutions for 20min, the OD 625 Colorimetric). According to the standard curve, the NH in the supernatant can be known 4 + concentration.
[0061] 2) Results
[00...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com