New method for performing data correction by using cell metabolite relative content as cell number index
A technology of cell metabolism and metabolomics data, which is applied in the field of metabolomics data correction and rapid quenching of biochemical metabolism of in vitro cultured cells, to achieve efficient extraction, ensure accuracy and reliability, and simplify experimental operations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
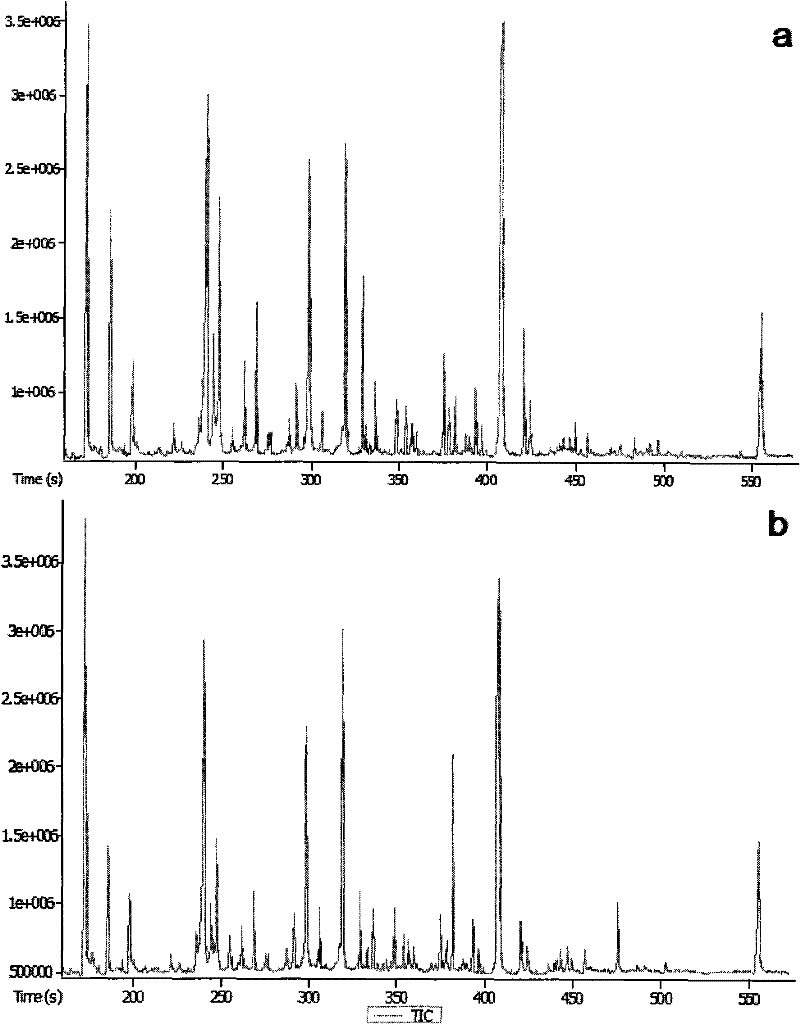
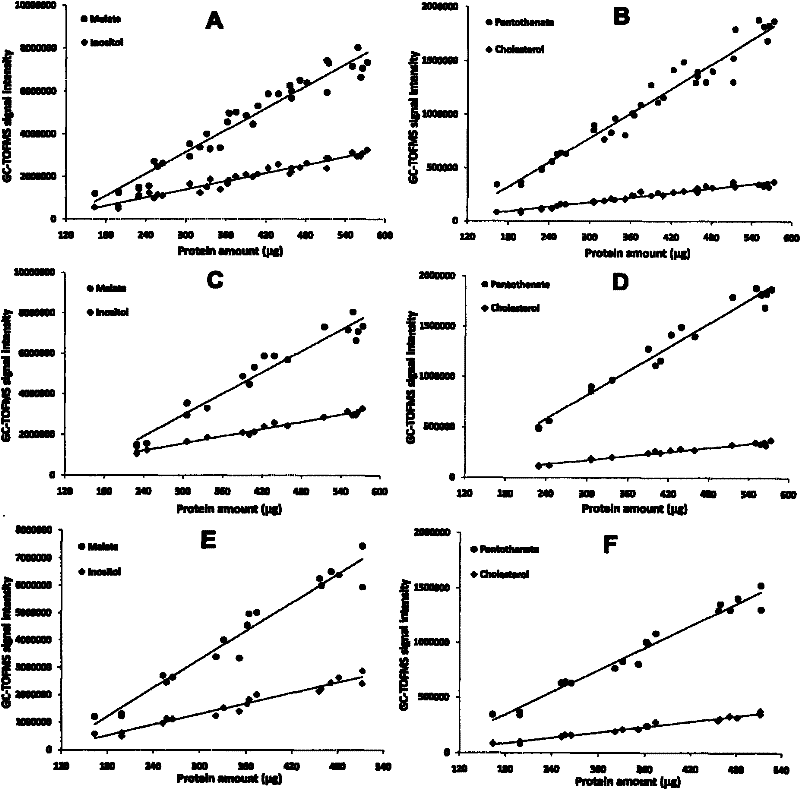
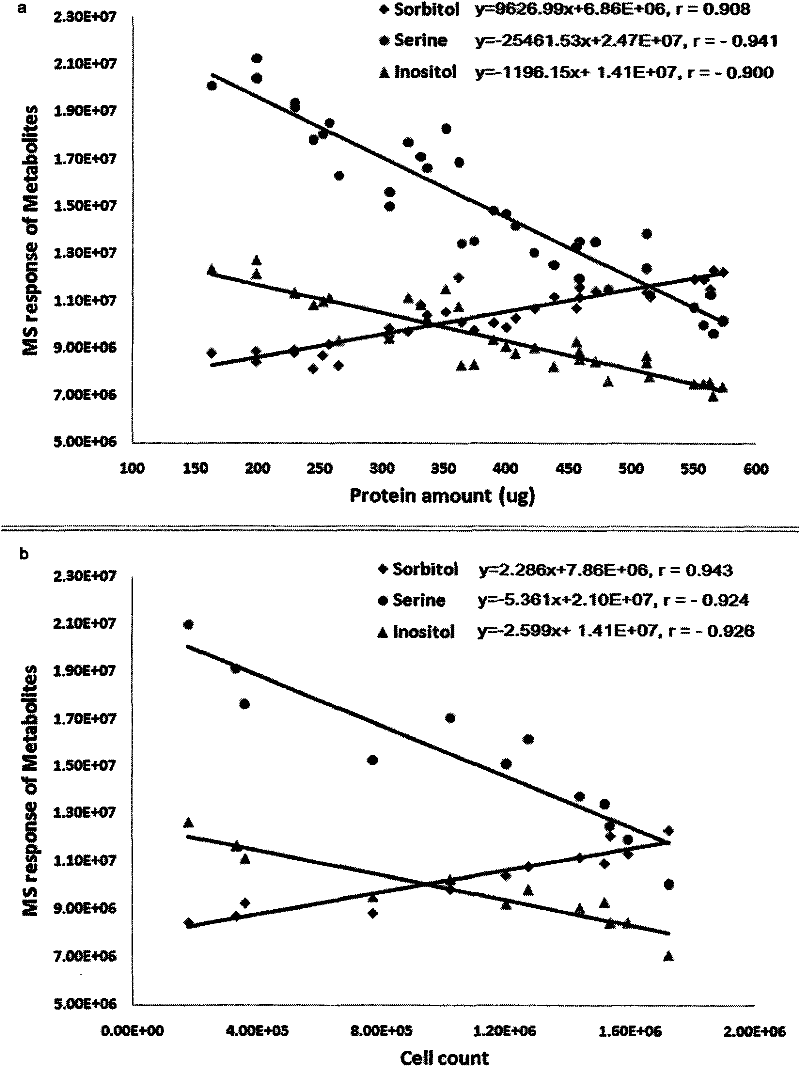
[0030] Cellular metabolites as indicators of cell number for correction of cellular metabolomics data - Madin-Darby canine kidney (MDCK) wild-type (MDCK-WT) and transfected human Mdr1 gene transfection Metabolomics Paradigm Study in Transfected (MDCK-MDR) Cells
[0031] 1. Experimental Protocol and Sample Collection
[0032] Serial volumes of cell suspension (A-F: 2.5, 2.0, 1.6, 1.2, 0.9, 0.6mL, MDCK-WT 600,000 / mL and MDCK-MDR 800,000 / mL) were seeded in six-well plates (n=4), and blanks were added The medium was such that the volume per space was 2.5 mL. The two cell lines were grown under the same culture conditions for about 3 days without changing the culture medium. The medium samples of the cells in each well were collected, and the cells were pretreated according to the above method, and 300 μL of ultrapure water was added to each well, and then frozen in a -70°C refrigerator for testing.
[0033] 2. Sample Processing
[0034] Cell sample: After repeated freezing and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com