Real-time fluorescence quantitative PCR method for detecting NPM1 genic mutation
A real-time fluorescence quantitative and fluorescent quantitative technology, which is applied in the direction of fluorescence/phosphorescence, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of many operation steps, easy pollution, and difficult detection ability to meet the requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1: Detection of NPM1 gene mutation in human fresh peripheral blood genomic DNA
[0029] Genomic DNA was extracted from the peripheral blood of 50 healthy people and 50 AML patients. For DNA extraction, DNA extraction can be performed using blood genomic DNA extraction kits provided by Qiagen, Promega, and Roche. Genomic DNA extraction can also be performed using the general DNA extraction method of the third edition of the Molecular Cloning Laboratory Guide. The following takes Qiagen's blood genome extraction kit operation steps as an example to illustrate the blood genome DNA extraction steps.
[0030] 1. Pipette 20 μl proteinase K into a 1.5 ml centrifuge tube.
[0031] 2. Add 200 μl of fresh blood.
[0032] 3. Add 200 μl Buffer AL, mix and shake for 15 sec.
[0033] 4. Incubate at 56°C for 10 minutes.
[0034] 5. Brief centrifugation.
[0035] 6. Add 200 μl absolute ethanol, mix and shake for 15 sec. Centrifuge briefly.
[0036] 7. Add the above mixt...
Embodiment 2
[0049] Embodiment 2: Detection of NPM1 mutant plasmids with different copy numbers
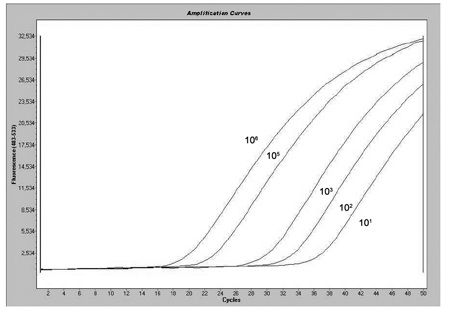
[0050] NPM1 was accurately quantified and diluted to 10 with TE buffer 6 Copies / 5 μl. to 10 6 Copy / 5μl sample was serially diluted to obtain 10 6 copies / 5μl sample, 10 5 copies / 5μl sample, 10 3 copies / 5μl sample, 10 2 copies / 5μl sample, 10 1 Copies / 5 μl sample.
[0051] Prepare the NPM1 mutant detection system according to Table 2, and perform PCR reaction.
[0052] After the reaction is completed, read the Ct value of the sample from the real-time fluorescent quantitative PCR instrument.
[0053] The result shows that the detection method of the present invention can detect 10 copies of the NPM1 gene mutation, the detection range is wide, and the linearization is good.
Embodiment 3
[0054] Example 3: Detection of NPM1 gene mutation from bone marrow tissue cDNA of AML patients
[0055] Bone marrow tissue samples were collected from 20 AML patients, and RNA was extracted using commercial total RNA extraction kits provided by Invitrogen and Qiagen. See the manufacturer's instructions for operating procedures.
[0056] The reverse transcription of RNA into cDNA can be carried out using a commercial kit, and this description uses Invitrogen’s Superscript II TM As an example, the main steps of reverse transcription will be described.
[0057] 1. Thaw all reagents on ice.
[0058] 2. Incubate 1 μg RNA at 70°C for 10 min, immediately place it on ice, and incubate for 5 min.
[0059] 3. Prepare the following reaction mixture
[0060] 5X RT Buffer 4μl
[0061] MgCl2 (50 mM) 2 μl
[0062] dNTP (10 mM each) 2 μl
[0063] DTT (100 mM) 2 μl
[0064] RNaseOut (40 U / Tl) 0.5μl
[0065] Random nonamer 5μl
[0066] Superscript II (200 U) 0.5μl
[0067] RNA 1-4μl ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com