Dedicated primer for auxiliarily detecting NDM-1 (New Metallo-Beta-Lactamase-1) gene and application thereof
A kind of NDM-1, auxiliary detection technology, applied in application, genetic engineering, plant genetic improvement and other directions, to achieve the effect of strong specificity, self-safety and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Embodiment 1, design and preparation of special primers
[0044] Each primer (FOP, BOP, FIP, BIP, FLP and BLP) shown in Table 1 was designed and synthesized.
[0045] The six primers shown in Table 1 form special primers and are used in Example 2.
[0046] Nucleotide sequence of table 1 primer
[0047]
[0048] The amino acid sequence of the NDM-1 protein is shown in sequence 7 of the sequence listing, its coding gene is shown in sequence 8 of the sequence listing, and the open reading frame is the 14th to 826th nucleotides from the 5' end of sequence 8 of the sequence listing.
Embodiment 2
[0049] Embodiment 2, application-specific primer detection NDM-1 gene
[0050] 1. Template preparation
[0051] The DNA (template) shown in the sequence 11 of the synthetic sequence list, in order to prevent the nucleotide sequence of the target sequence from being translated into an active functional protein, individual nucleotides in the target sequence were replaced, in order to facilitate the integration with For other template discrimination, the DNA shown in Sequence 11 is also called NDM-1 gene.
[0052] 2. Application of special primers to detect NDM-1 gene
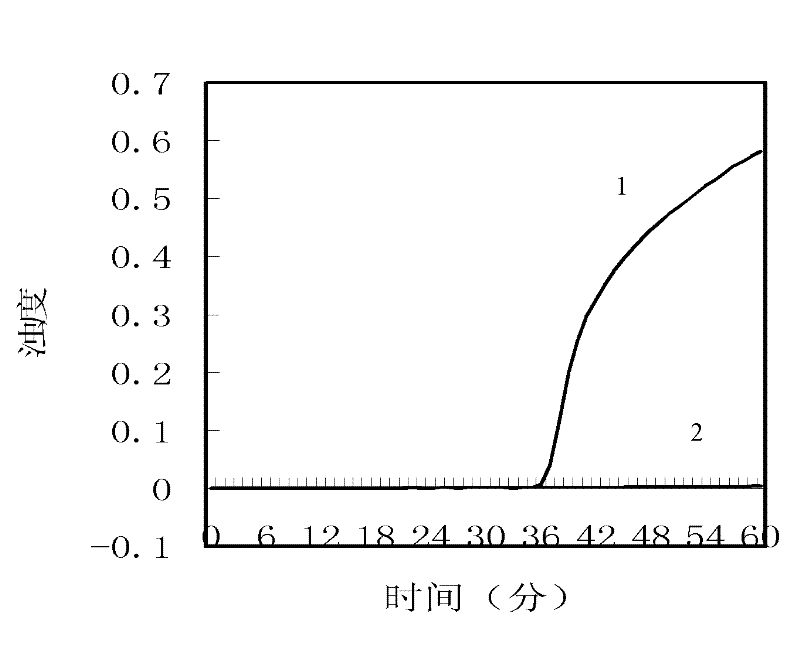
[0053] 1. Interpret the results by observing the change of fluorescent color (fluorescence colorimetric method)
[0054] The template synthesized in Step 1 (concentration: 2.9 ng / μl) was used for LAMP (loop-mediated constant temperature amplification). The LAMP reaction system is shown in Table 2.
[0055] Table 2 LAMP reaction system (25μL)
[0056] components
The final concentration in the react...
Embodiment 3
[0085] Embodiment 3, the sensitive determination of special primer
[0086] 1. Prepare the template
[0087] The NDM-1 gene shown in the sequence 11 of the sequence listing was synthesized and used as a template after gradient dilution. The concentrations of the templates were as follows: 2.9×10 -6 ng / μl, 2.9×10 -7 ng / μl, 2.9×10 -8 ng / μl.
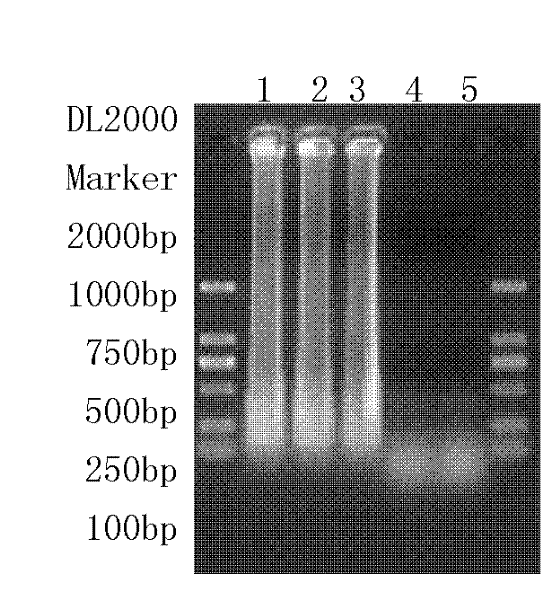
[0088] Second, the sensitivity of electrophoresis detection
[0089] The templates synthesized in Step 1 were used to carry out LAMP (loop-mediated constant temperature amplification), and the LAMP reaction system is shown in Table 3.
[0090] Use 2 μL of double distilled water as the template blank control, 2 μL of Klebsiella pneumoniae genomic DNA (82.8ng / μl) as negative control 1, and 2 μL of Escherichia coli genomic DNA (52.4ng / μl) as negative control 1 Negative control 2.
[0091] Mix the reagents in Table 3 except the template for use; pre-denature the template at 96°C for 3 minutes; then mix the template with other well-mixed ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com