Method for extracting nucleuses of mature leaves of fruit trees suitable for flow cytometry analysis
A technology of flow cytometry and mature leaves, which is applied in the field of cell biology, can solve the problems of flow cytometry limitations and other problems, achieve good detection results, little impact, and maintain the integrity of the effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
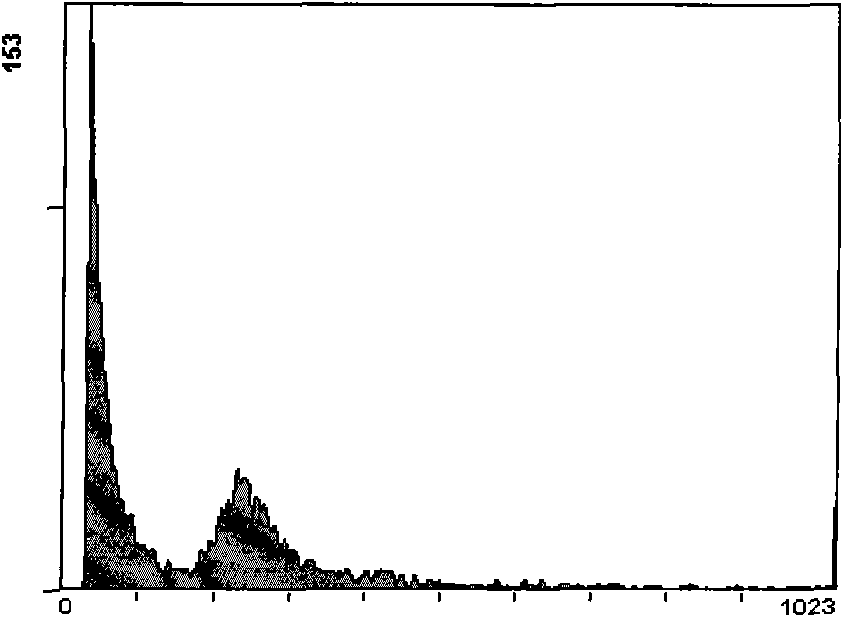
[0030] Take 200mg of mature leaves of Fuji apple in the field, put them into a pre-cooled petri dish, first treat with 5% sodium hypochlorite for 10 minutes, then add 3ml of pre-cooled ether, pour off the ether immediately after 30 seconds, and quickly wash with cold extraction buffer 5 times, grind in a mortar with 2 mL of nucleus extraction buffer, filter through a 500-mesh screen, centrifuge the filtrate at 4°C (2000 rpm) for 5 minutes, rinse 3 times, and collect deposited nuclei. Suspended with 400 μl of staining buffer, placed in a dark place at room temperature, stained for 30 min, and then tested on the machine.
[0031] The components of the cell nucleus extracting solution are: 15mmol / L Tris, 40mmol / L KCl, 20mmol / L NaCl, 5mmol / L Na 2 EDTA, 10mmol / L MgSO 4 , 50mmol / L sucrose and 1.0% (v / v) Triton X-100, add ddH by volume 2 0 to 200ml, after adjusting the pH to 7.5, add 200 μl of 15mmol / L mercaptoethanol.
[0032] The staining buffer used was to add 20 mg / L RNase A a...
Embodiment 2
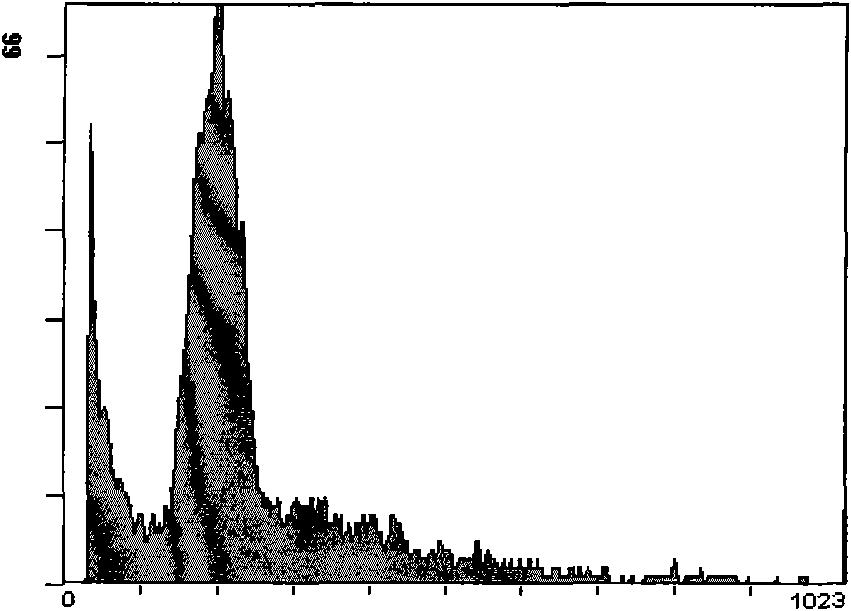
[0038] Mix 200 mg of mature leaves of Muscat (2X) and Kyoho (4X) grapes in the field, first treat with 8% sodium hypochlorite for 15 minutes, then put it into a pre-cooled petri dish, add 3ml of pre-cooled ether, pour it out immediately after 40 seconds Ethyl ether was quickly washed 3 times with cold extraction buffer, ground in a mortar with 1 mL of extraction buffer, filtered through a 600-mesh sieve, and the filtrate was centrifuged (2000 rpm) at 4 °C for 5 minutes, rinsed 3 times, and collected deposited nuclei. Suspended with 500 μl of staining buffer, placed in a dark place at room temperature, stained for 15 minutes, and then tested on the machine.
[0039] The components of the cell nucleus extracting solution are: 20mmol / L Tris, 60mmol / L KCl, 10mmol / L NaCl, 2mmol / L Na 2 EDTA, 20mmol / L MgSO 4 , 40mmol / L sucrose and 0.5% (v / v) Triton X-100, add ddH by volume 2 O (deionized water) to 200ml, after adjusting the pH to 7.8, add 200μl of 15mmol / L mercaptoethanol.
[004...
Embodiment 3
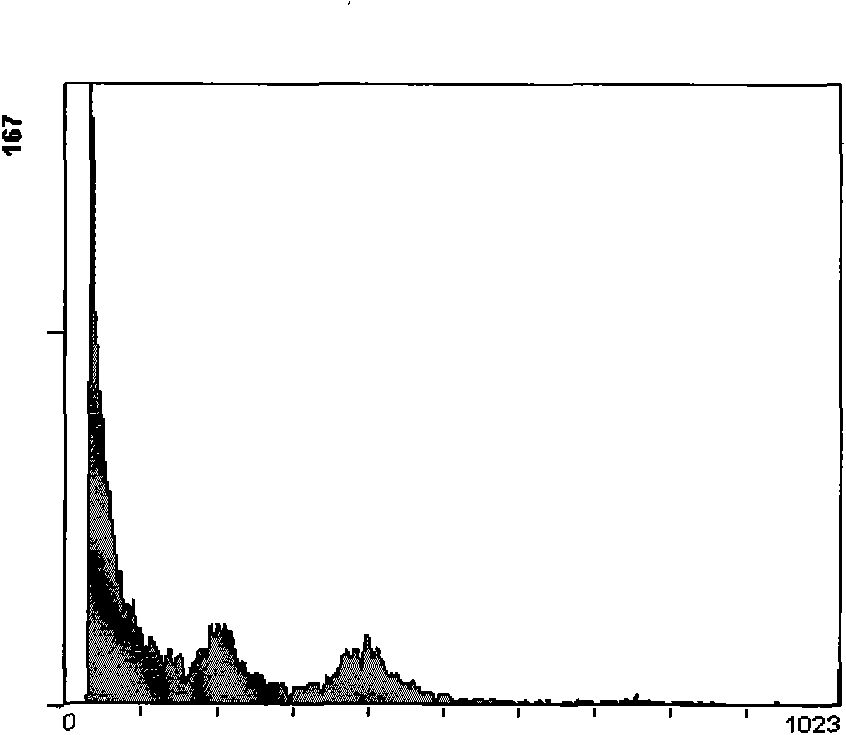
[0046] Take 200 mg of pineapple leaves in the field, first treat them with 10% sodium hypochlorite for 10 minutes, then put them into a pre-cooled petri dish, add 3ml of pre-cooled ether, pour off the ether immediately after 50 seconds, and quickly wash 3 times with cold extraction buffer , put into a mortar with 2ml of extraction buffer and grind, filter through a 400-mesh sieve, centrifuge the filtrate at 2°C (1500r / min) for 10 minutes, rinse 3 times, and collect the deposited nuclei. Suspended with 600 μl of staining buffer, placed in a dark place at room temperature, stained for 30 minutes, and then tested on the machine. Such as Figure 3a , Figure 3b shown.
[0047] The components of the cell nucleus extracting solution are: 20mmol / L Tris, 40mmol / L KCl, 10mmol / L NaCl, 5mmol / L Na 2 EDTA, 20mmol / L MgSO 4 , 50mmol / L sucrose and 1.0% (v / v) Triton X-100, add ddH by volume 2 O (deionized water) to 200ml, after adjusting the pH to 8.0, add 200μl of 15mmol / L mercaptoethano...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com