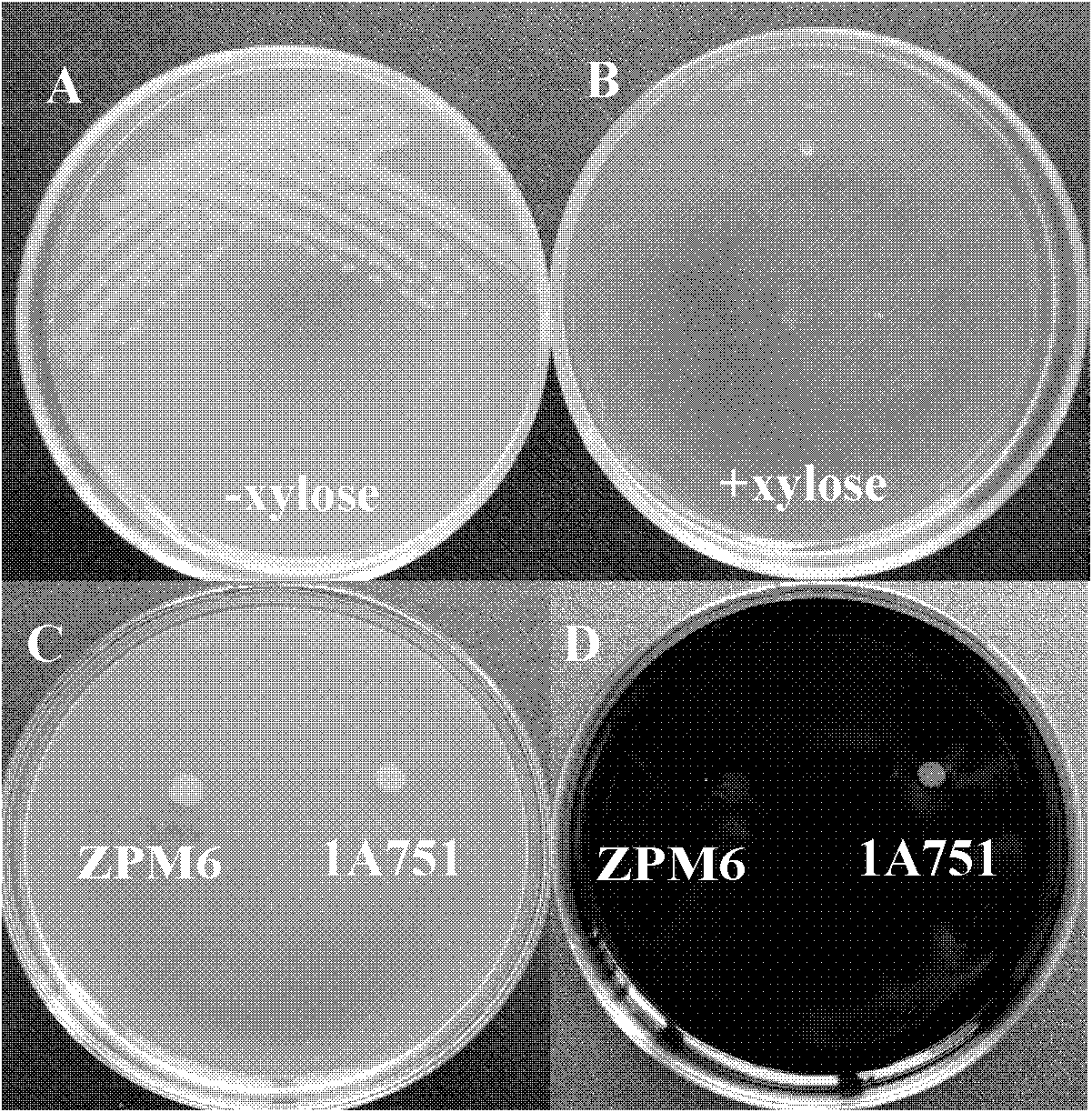Expression cassette mazF-cassette for expressing escherichia coli toxin protein mazF gene and construction method and application thereof
A technology for expressing Escherichia coli and toxin proteins, applied in the field of genetic engineering, can solve the problems of long cycle, affecting the expression of adjacent genes, and adverse effects on the physiological state of strains
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
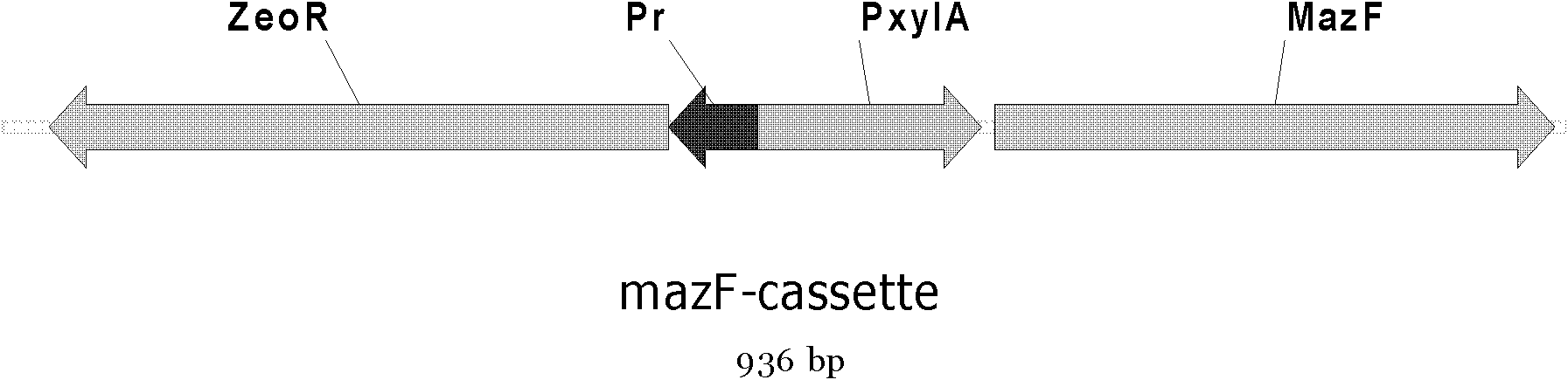
[0070] The construction of the expression cassette of embodiment 1 Escherichia coli toxin protein mazF gene
[0071] (1) Primers:
[0072] 5ZeoRF: ggcGTCGACGGATCCGAATTCAAGCTTCAGTCCTGCTCCTCGGCCAC
[0073] 3ZeoRR: TTCATGAAAGACTTGATATGGCTTTTTATATGTG
[0074] 5PxylF: CATATCAAGTCTTTCATGAAAAACTAAAAAAATATT
[0075]3PxylR:ACCAGATCCTCCTTTAGATGCATTTTATGTCATATTGTA
[0076] 5SOEmazF;CATCTAAAGGAGGATCTGGTAATGGTAAGCCGATA
[0077] 3SOEmazR: ggcTCTAGACTACCCCAATCAGTACGTTAAT
[0078] (2) Gene amplification
[0079] The zeocin resistance gene was amplified by PCR using the plasmid pDGCZ as a template and 5ZeoRF / 3ZeoRR as primers; the xylose promoter gene was amplified by PCR using the plasmid pSG1729 as a template and 5PxylF / 3PxylR as primers; PCR amplification of the mazF gene for primers;
[0080] The reaction systems are respectively: 5×PrimeSTAR Buffer (Mg 2+ ) 10 μl, dNTP Mixture (2.5 mM each) 4 μl, upstream primer 1 μl, downstream primer 1 μl, template DNA 2 μl, PrimeSTAR HS DNA Pol...
Embodiment 2
[0094] Example 2 Verification of the expression cassette function of the Escherichia coli toxin protein mazF gene
[0095] (1) Verification of amylase activity
[0096] The strains verified as positive transformants by PCR, named ZPM6 (Zeocin-PxylA-MazF) and the control strain 1A751 were respectively planted on solid plates of LB+1% starch, and counterstained with iodine solution the next day to detect their Amylase activity ( figure 2 C and D); the results showed that the ZPM6 strain could not form a transparent image on starch culture, and lost amylase decomposing bacteria, indicating that mazF-cassette was inserted into the amylase gene (amyE) position; while the control strain 1A751 was due to the presence of complete starch The enzyme gene (amyE) can form a transparent image on starch medium.
[0097] (2) Verification of reverse screening function
[0098] The above-mentioned ZPM6 (Zeocin-PxylA-MazF) and the control strain 1A751 were respectively streaked on solid pla...
Embodiment 3
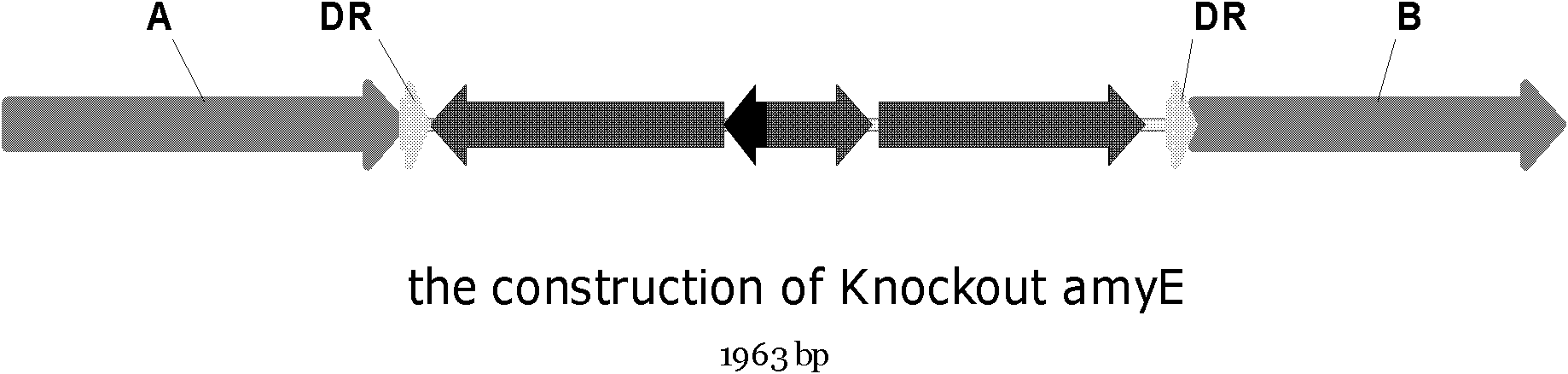
[0099] Embodiment 3 Bacillus subtilis amylase gene (amyE) does not have the deletion of resistance marker
[0100] (1) Primer
[0101] 5maZF (Box): AATCAATAATGGACCAGACGACAGTCCTGCTCCTCGGCCAC
[0102] 3SO EmazR: GGCTCTAGACTACCCCAATCAGTACGTTAAT
[0103] 5fPamyE: AGTCTTCAAAAAATCAAATAAGGAGT
[0104] 3fPamyE: TCGTCTGGTCCATTATTGATTTGATAAACG CTTAACCTCATTGGAAATCGCG
[0105] 5bPamyE: TGATTGGGTAGTCTAGAGCCCAGATGCGAATACAACAAAAGC
[0106] 3bPamyE: GTAAGTCCCGTCTAGCCTTGCCCTC
[0107] Note: The underline is the 30bp of the direct repeat (Direct-repeat) with the homologous fragment at the 3' end.
[0108] (2) Gene amplification
[0109] The mazF-cassette gene was amplified by PCR using the Bacillus subtilis ZPM6 genome as a template and 5maZF(Box) / 3SOEmazR as primers; using the Bacillus subtilis 1A751 genome as a template and 5fPamyE / 3fPamyE as primers to amplify the 5' homologous fragment by PCR; The genome of Bacillus subtilis 1A751 was used as a template, and 5bPamyE / 3bPamyE was use...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com