Fluorescence PCR (polymerase chain reaction) kit for detecting yersinia enterocolitica
A technology for enterocolitis and Yersinia, which is applied in the field of real-time fluorescent PCR reagents for detecting Yersinia enterocolitica, can solve problems such as no specific research content, and achieves a simple and fast detection process, high sensitivity, Avoid tedious effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
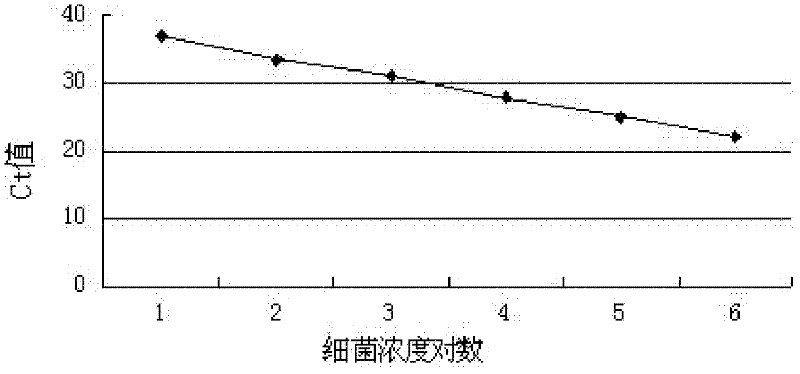
Image
Examples
Embodiment 1
[0041] In the Yersinia enterocolitica fluorescent PCR kit of the present invention, the reagents in the kit include a front primer with a nucleic acid sequence of 5'-AATGCTGTCTTCATTTGGAGC-3', a back primer with a nucleic acid sequence of 5'-ATCCCAATCACTACTGACTTC-3', TaqMan probe whose nucleic acid sequence is 5'-CAAGCAAGCTTGTGATCCTCCG-3', PCR amplification buffer, mixed liquid containing DNA polymerase, magnesium ions and four kinds of deoxynucleosides dATP, dCTP, dGTP, dTTP, deionized water and bacteria body concentration of 10 7 Positive standard solution of cfu / ml. The 5'-end label of the TaqMan probe is 6-carboxysuccinimide ester, and the 3'-end label is 6-carboxytetramethylrhodamine; wherein the molar ratio of the front primer, the back primer, and the TaqMan probe 1:1:1; the molar ratio of the four dNTPs is dATP:dCTP:dGTP:dTTP=l:l:l:l. This is the fluorescent PCR kit for detecting Yersinia enterocolitica of the present invention.
Embodiment 2
[0042] Example 2: Quantitative detection of Yersinia enterocolitica in stool samples
[0043] 1. Bacterial DNA Extraction
[0044] (1) Add phosphate buffered saline (PBS) 4 times the amount of the sample and mix well with the stool sample, bathe in 100°C for 15 minutes, centrifuge at 8000 rpm (revolutions per minute) for 5 minutes, take 500 μL of supernatant, add 50 μL of 10% ten Sodium dialkyl sulfate (SDS), 20 μL proteinase K, 50 ° C water bath for 1 h, shake once every 15 min.
[0045] (2) Add an equal amount of phenol, shake well, centrifuge at 10000rpm for 5min.
[0046] (3) Take the supernatant, add 300 μL each of phenol and chloroform, shake well, and centrifuge at 10,000 rpm for 5 minutes.
[0047] (4) Take the supernatant (do not inhale sundries), add an equal amount of chloroform to mix, centrifuge at 10000rpm for 5min.
[0048] (5) Take the supernatant (do not inhale sundries), add 1000 μL of frozen absolute ethanol, add 1 / 10 volume, 3 mol / L NaAc, and set it at -...
Embodiment 3
[0061] Example 3: Quantitative detection of Yersinia enterocolitica in food samples
[0062] 1. Bacterial DNA Extraction
[0063] (1) Add phosphate buffer saline (PBS) 4 times the amount of the sample and grind and homogenize the food sample, bathe at 100°C for 15 minutes, centrifuge at 8000 rpm (revolutions per minute) for 5 minutes, take 500 μL of supernatant, add 50 μL of 10% ten Sodium dialkyl sulfate (SDS), 20 μL proteinase K, 50 ° C water bath for 1 h, shake once every 15 min.
[0064] (2) Add an equal amount of phenol, shake well, centrifuge at 10000rpm for 5min.
[0065] (3) Take the supernatant, add 300 μL each of phenol and chloroform, shake well, and centrifuge at 10,000 rpm for 5 minutes.
[0066] (4) Take the supernatant (do not inhale sundries), add an equal amount of chloroform to mix, centrifuge at 10000rpm for 5min.
[0067] (5) Take the supernatant (do not inhale sundries), add 1000 μL of frozen absolute ethanol, add 1 / 10 volume, 3 mol / L NaAc, and set it a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com