Detection method for slow virus infected cell
A detection method and a technology for infecting cells are applied in the field of detection of lentivirus-infected cells, and can solve problems such as troublesome detection of lentivirus-infected cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
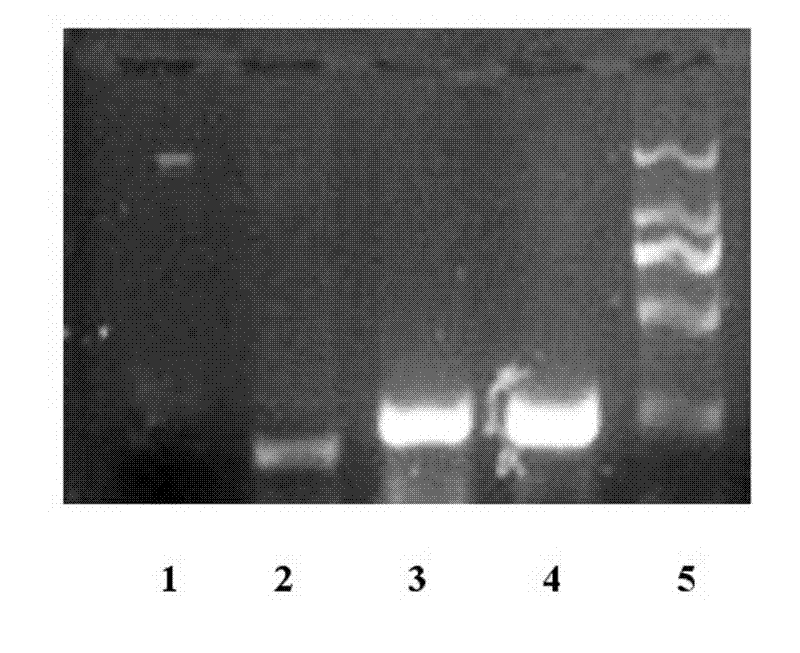
[0018] The detection method of lentivirus-infected cells of the present invention, its steps are:
[0019] 1) Synthetic random DNA ligand library:
[0020] The screening of oligonucleotide ligands requires the construction of artificially synthesized single-stranded random oligonucleotide libraries, in which the random sequence length is 20-40 bases, and the library capacity is 10E^14~10E^15 (10 out of 14~15 power) between. Because single-stranded random oligonucleotide fragments, especially single-stranded RNA, are prone to form secondary structures such as hairpins, pockets, pseudosections, and G-tetramers, they can interact with proteins, nucleic acids, oligopeptides, amino acids, organic substances, and even metal ions Combined to form a complex with strong binding force.
[0021] Such as -TGTTGTGAGCCTCCT- N25 (25 random DNA sequences), -TTATTCTTGTCTCCC- is one of them.
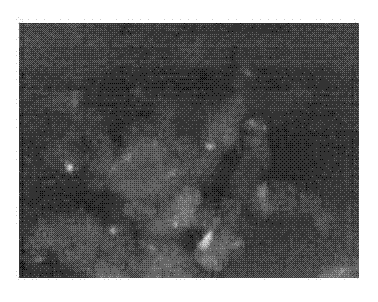
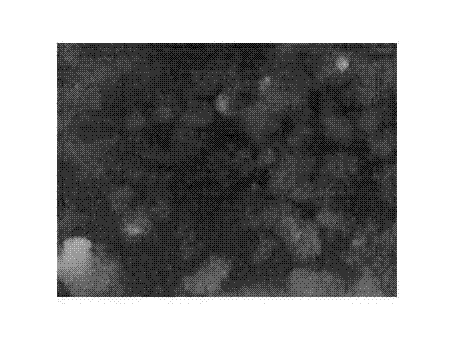
[0022] 2) Lentivirus infection of HEK293 cells
[0023] This protocol uses two lentiviruses to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com