Method for quickly identifying bloodstream infection pathogenic bacteria
An identification method, a technology of pathogenic bacteria, applied in the field of microbial inspection, can solve the problems of unfavorable popularization and high price
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
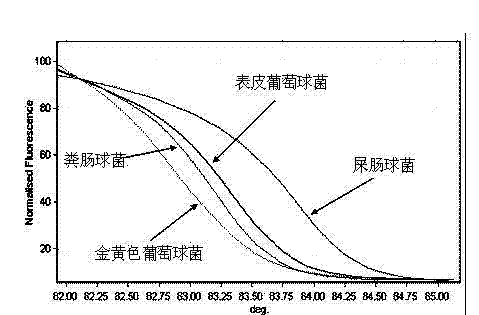
[0026] The identification of embodiment one positive cocci:
[0027] Gram-positive cocci on the smear;
[0028] Amplify the 16S gene fragment with 16SBF and 16SBR primers;
[0029] High-resolution melting curve analysis directly identifies species.
[0030] 20 cases of blood culture positive specimens were tested, among them, 7, 11, 16, and 18 were positive cocci. High-resolution melting curve analysis identified 2 cases of Staphylococcus aureus, 1 case of Staphylococcus epidermidis, and 1 case of Enterococcus faecalis infection.
[0031]
Embodiment 2
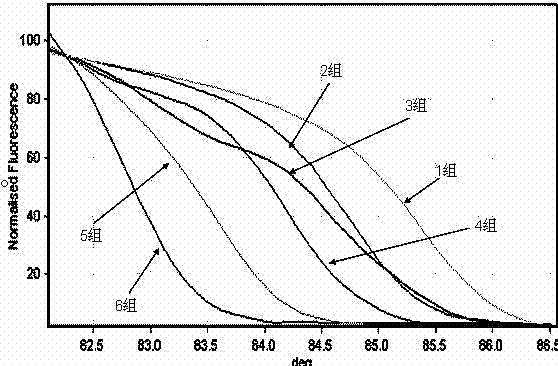
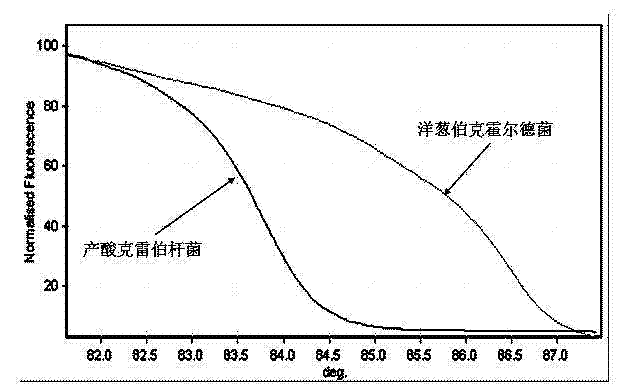
[0032] The identification of embodiment two negative bacilli:
[0033] smear Gram-negative bacilli;
[0034] The 16S gene fragment was amplified with 16SAF and 16SBF primers.
[0035] The common negative bacilli of 12 kinds of bacteria can be divided into 6 groups, the first group is Escherichia coli, the second group is Pseudomonas aeruginosa, Serratia marcescens, Stenotrophomonas maltophilia, and the third group is producing Enterobacter aerogenes, Klebsiella pneumoniae, Enterobacter cloacae, Klebsiella oxytoca, Burkholderia cepacia in group 4, Acinetobacter lophi in group 5, and Baumannii in group 6 Acinetobacter.
[0036] The determination of 30 cases of positive blood culture samples was completely consistent with the results obtained by subsequent traditional methods. It is confirmed that the rapid identification of pathogenic bacteria of bloodstream infection in the present invention requires only 3 hours from blood culture reporting positive, PCR amplification, and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com