Aptamer sequence of hepatitis B virus (HBV) core antigen and application of nucleic aptamer sequence
A nucleic acid aptamer and core antigen technology, which can be used in antiviral agents, pharmaceutical formulations, microbial measurement/inspection, etc., and can solve the problems of nucleic acid aptamers, reports or publications of hepatitis B virus.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Embodiment 1: HBc gene cloning
[0026] Using known molecular cloning techniques, based on the recombinant plasmid pBS_HBV3.6II containing double copies of the HBV genome sequence, primers were designed according to the HBc gene sequence:
[0027] HBc_L: 5'-GCCCATATGGACATTGACCCGTA-3'
[0028] HBc_R: 5'-GCCCTCGAGTCAAAACAACAGTAGTTT-3'
[0029] Configure PCR system (total volume 50ul): H 2 O 38ul, 10×PCR buffer 5ul, 25mM MgCl 2 3ul, dNTP(10mM) 1ul, primer mix 1.5ul (H 2 O: 100uM HBc_L: 100uM HBc_R=4:0.5:0.5), DNA template 1ul, PrimeStar enzyme 0.5ul. Amplify on a PCR machine under the following conditions: heat pre-denaturation at 94°C for 2 min, denature at 94°C for 30 s, anneal at 55°C for 30 s, extend at 72°C for 20 s, 30 cycles, extend at 72°C for 8 min, and hold at 4°C for <1 h.
[0030] After the reaction, 5 ul of the product was electrophoresed in 1% agarose gel at 100V constant voltage, and after identifying and amplifying a DNA fragment consistent with the...
Embodiment 2
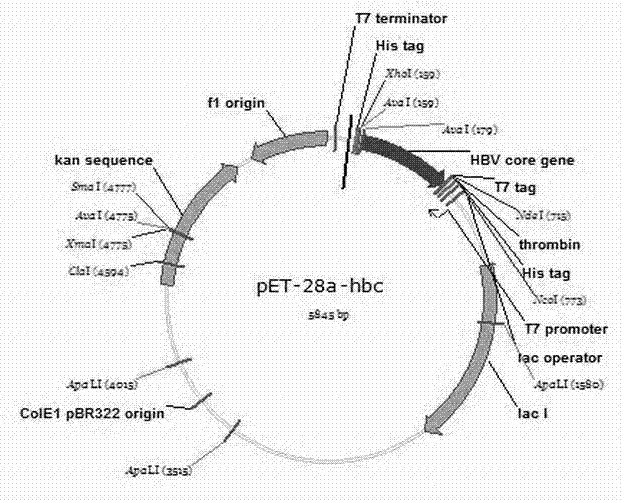
[0033] Embodiment 2: HBc expression and purification
[0034] A single colony of Escherichia coli containing the recombinant expression plasmid pET28a-HBc was inoculated into 5 mL of LB medium containing Kan antibiotics, and cultured overnight at 37°C. Transfer the overnight culture to 200 mL of fresh medium according to the volume ratio of 1:100, culture for 2-4 hours, and wait until OD 600 When the value reaches 0.6, add IPTG (final concentration is 0.1mM), induce culture at 30°C for 8h. The cells were collected by centrifugation at 4000rpm for 30min.
[0035] Resuspend the bacteria with 5mL lysate (50mM Tris-HCl (pH8.5~9.0), 2mM EDTA, 100mM NaCl, 0.5% Triton X-100, 1mg / ml lysozyme), and divide into three 2ml tubes, -80 degrees for 30 minutes. 100w ultrasonic crushing in ice bath (2s~5s, 60~80 times). Mix 1ml supernatant with 500ul Ni-NTA Agarose beads, add to the purification column, wash with 2ml PBS+Mg (1mM MgCl 2 ) after washing twice, resuspend with 2ml PBS+Mg an...
Embodiment 3
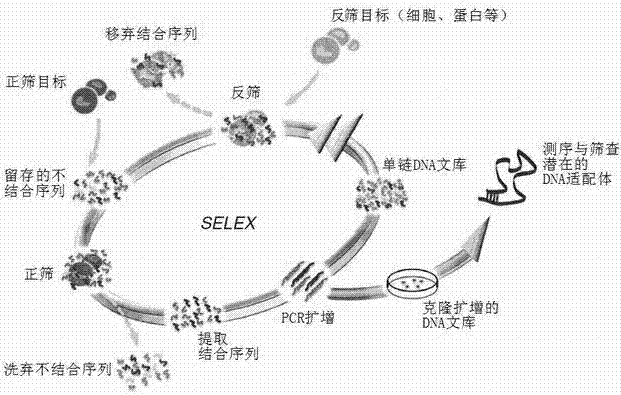
[0038] Example 3: Nucleic acid aptamer screening (one round)
[0039] First round oligo DNA library sequence: 5'-acgctcggatgccactacagNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNctcatggacgtgctggtgac-3';
[0040] Primers used for enrichment:
[0041] Aptamer_L: 5'-FAM-acgctcggatgccactacag-3';
[0042] Aptamer_R: 5'-biotin-gtcaccagcacgtccatgag-3';
[0043] Determination of OD of oligomeric DNA library 260 Afterwards, it was centrifuged and dried. Dissolve the library with 300ul PBS+Mg, take 200pmol (2.5nmol in the first round), put it on ice at 95°C for 8min, and then centrifuge quickly.
[0044] Reverse screening: Aspirate 200pmol of the library into the reverse screening target equivalent to 40pmol of the positive screening target, and make up to 800ul with PBS+Mg. Shake at 37°C for 30 minutes. Inhale the Milipore ultrafiltration tube for quick centrifugation (<1000rpm), and take the filtrate.
[0045] Positive sieve: Take 40pmol positive sieve target, centrifuge at 800rpm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com