Method for detecting gene mutation genotyping based on Taqman-ARMS (Amplification Refractory Mutation System) technology and kit
A technology for technical detection and kits, applied in the fields of biotechnology and medicine, can solve the problems of easy contamination sensitivity, high cost, complicated procedures, etc., and achieve the effects of simple and fast operation, high safety, and pollution reduction.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] The human EGFR gene is located on chromosome 7 of the genome, the EGFR genomic DNA (Genebank: NG_007726.3), and the full-length cDNA encoding the EGFR gene is shown in SEQ ID No: 1. According to the missense mutation of EGFR gene mainly concentrated in exon 21, the sequence of exon 21 of EGFR gene is shown in SEQ ID No: 2, the mutation site mainly has 2573 missense mutations, that is, the 2573 position is changed from T to G ( Abbreviated as L858R).
[0043] According to the L858R mutation type, use Primer 5 to design ARMS primer pairs, make the missense mutation site at the last position of the 3' end of the upstream primer, and introduce mismatch mutation bases at the 1st, 2nd and 3rd positions upstream of the mutation site , the specific primers are shown in Table 1, and the downstream primer is: 5'-aaacaatacagctagtgggaaggc-3' (SEQ ID No: 3)
[0044] Table 1. ARMS upstream primer sequences
[0045] 3' mismatch position Upstream primer sequence (5'→3') ...
Embodiment 2
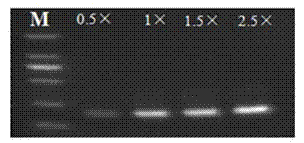
[0050] With the nucleotide sequences of SEQ ID No: 5 and SEQ ID No: 3 as primers, the positive control prepared in Example 1 was used as a template to carry out PCR, and the PCR buffer concentrations in the PCR reaction were 0.5×, 1×, 1.5×, 2× and 2.5×, the concentration of other components is the same (the final concentration of each component in the 20μL system is 0.25mM dNTP, 2.5mM MgCl 2 , 1U / reaction DNA polymerase, 250nM primers, 30ng template DNA and the rest water). The PCR reaction conditions were: pre-denaturation at 95°C for 5 minutes, one cycle; 40 cycles of amplification, deformation at 95°C for 20 seconds, annealing and extension at 60°C for 40 seconds; and finally cooling at 40°C for 15 seconds. The PCR amplification products were identified by electrophoresis, and the results were as follows: figure 2 shown. Depend on figure 2 It can be seen that the bands were amplified in the range of PCR buffer concentration gradient from 0.5× to 2.5×, and the result wa...
Embodiment 3
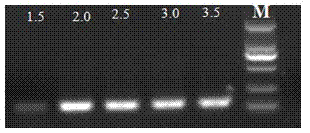
[0052] With the nucleotide sequences of SEQ ID No: 5 and SEQ ID No: 3 as primers and the positive control prepared in Example 1 as a template, MgCl 2 For PCR with different concentrations, the conditions of other components are the same (the final concentration of each component in the 20μL system is 1×PCR buffer, 0.25mM dNTP, 1U / reaction DNA polymerase, 250nM primer, 30ng template DNA, and the balance is water), MgCl 2 The concentrations were 1.5mM, 2.0mM, 2.5mM, 3.0mM and 3.5mM. The PCR reaction conditions are the same as in Example 2, and the amplified product is identified by electrophoresis, and the results are as follows: image 3 shown. Depend on image 3 It can be seen that in MgCl 2 The concentration in the range of 1.5mM~3.5mM can amplify the target band, when MgCl 2 It works best at a concentration of 2.0-2.5mM.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com