Re-engineering mediated site-directed mutagenesis method
A site-directed mutagenesis and recombinase technology, applied in the field of genetic engineering, can solve the problems of inability to carry out extension, cumbersome operation, long time-consuming, etc., achieving good economic prospects and simplifying the operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1. Construction of nanA expression vector
[0043] Design primer AL1: 5'-GGG GGATCC ATGGCAACGAATTTACGTGGC-3', (SEQ ID NO. 1); AL2: 5'-GGG AAGCTT TCACCCGCGCTCTTGCATCAAC-3', (SEQ ID NO.2), respectively introduced BamHI and HindIII restriction sites are underlined. Using the genomic DNA of the original strain of Escherichia coli MG1655 as a template, the 0.9kb nanA gene was amplified from AL1-AL2, and the 0.9kb was digested with BamHI-HindIII and cloned into the BamHI-HindIII site of pET30a(+) to obtain E. coli aldolization Enzyme expression vector pLS2042.
Embodiment 2
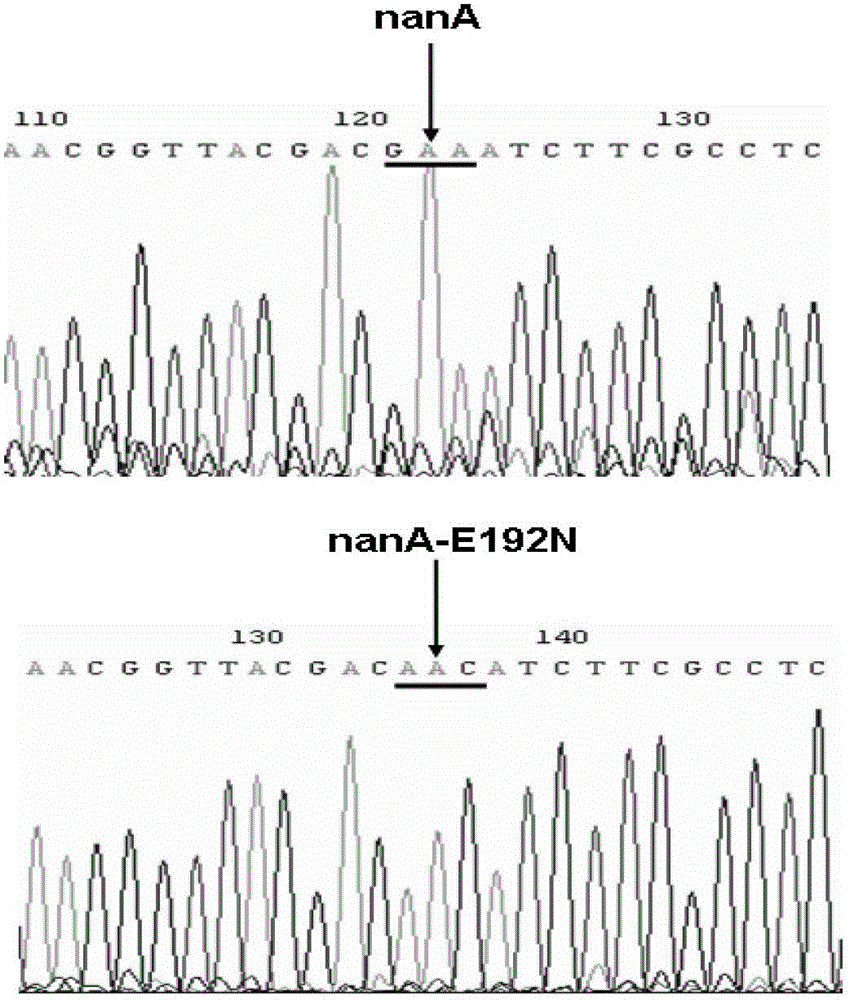
[0044] Example 2 Site-directed mutation of nanA gene E192N
[0045] 1. Preparation and transformation of E. coli competent cells expressing the Red recombinase gene
[0046] Transform pLS2042 into LS-GR, and select with 25 μg / ml gentamycin and 30 μg / ml kanamycin to obtain strain LS-GR / pLS2042. Inoculate the single bacterium colony of gained bacterial strain to 3ml in the LB liquid culture medium that contains the gentamycin of 25 μ g / ml and the kanamycin of 30 μ g / ml, 37 o C Shake overnight, transfer 1 / 50 volume to 50 ml LB liquid medium of the same resistance, and shake to culture the cells to OD 600 At about 0.2, add L-arabinose at a final concentration of 0.2% to induce recombinase, and continue to culture until OD 600 About 0.4, pour the bacterial solution into a pre-cooled centrifuge tube, ice bath for 10 minutes, 4 o C, centrifuge at 5000rpm for 5 minutes, discard the supernatant. Wash the pellet twice with ice-cold 10% glycerol, and finally suspend in 200...
Embodiment 3
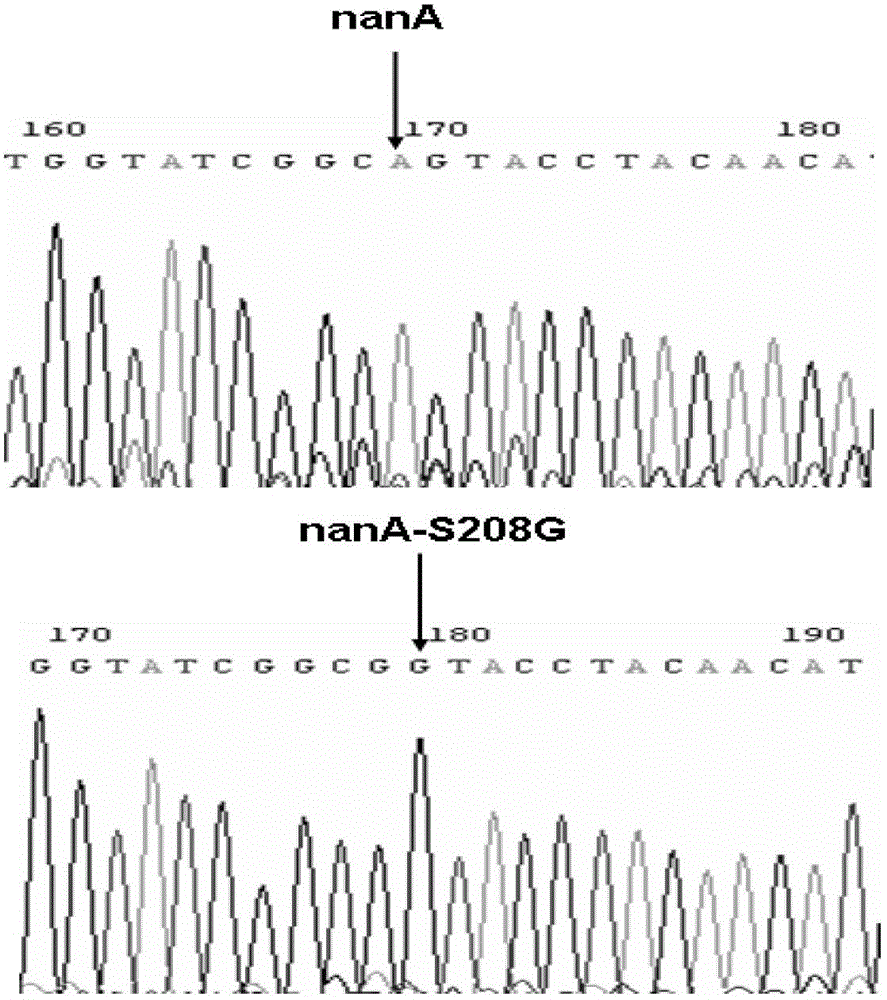
[0057] Example 3. Site-directed mutation of nanA gene S208G
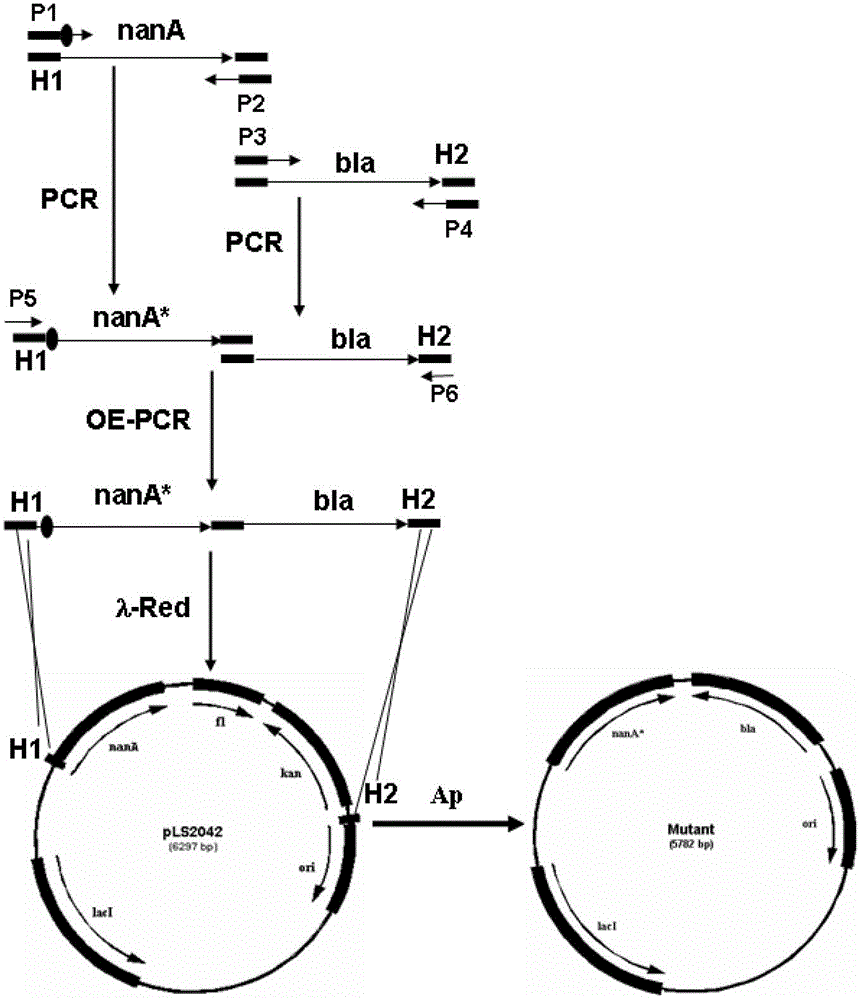
[0058] Design primer R555: 5'-cgaaatcttcgcctctggtctgctggcgggcgctgatggtggtatcggc G GTACCTACAACATCATGGGCTG -3', (SEQ ID NO.10), in this primer, the 50bp homology arm upstream of the target site is indicated by lowercase letters, the mutation site is indicated by italic letters, and the amplification primer is indicated by underlined letters; R563:5 '-ACGAAATCTTCGCCTCTGGTC-3', (SEQ ID NO.11), R563 is the sequence of about 21 bases at the front of R555. The primer R552 for amplifying the nanA gene fragment, the primers R553 and R554 for amplifying bla, the downstream short amplification primer R561 and the sequencing primer R566 are all the same as those in the E192N facility example. R555 and R552 respectively correspond to figure 1 P1 and P2 in, R553 and R554 respectively correspond to figure 1 P3 and P4, R56 and R562 respectively correspond to figure 1 P5 and P6 in.
[0059] Using pLS2042 as a templ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com