Bacterial surface display and screening of thioether-bridge-containing peptides
A thioether, fusion peptide technology, applied in the direction of peptides, fusion peptides, chemical instruments and methods, etc., can solve problems such as difficulty in introducing synthetic peptides
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
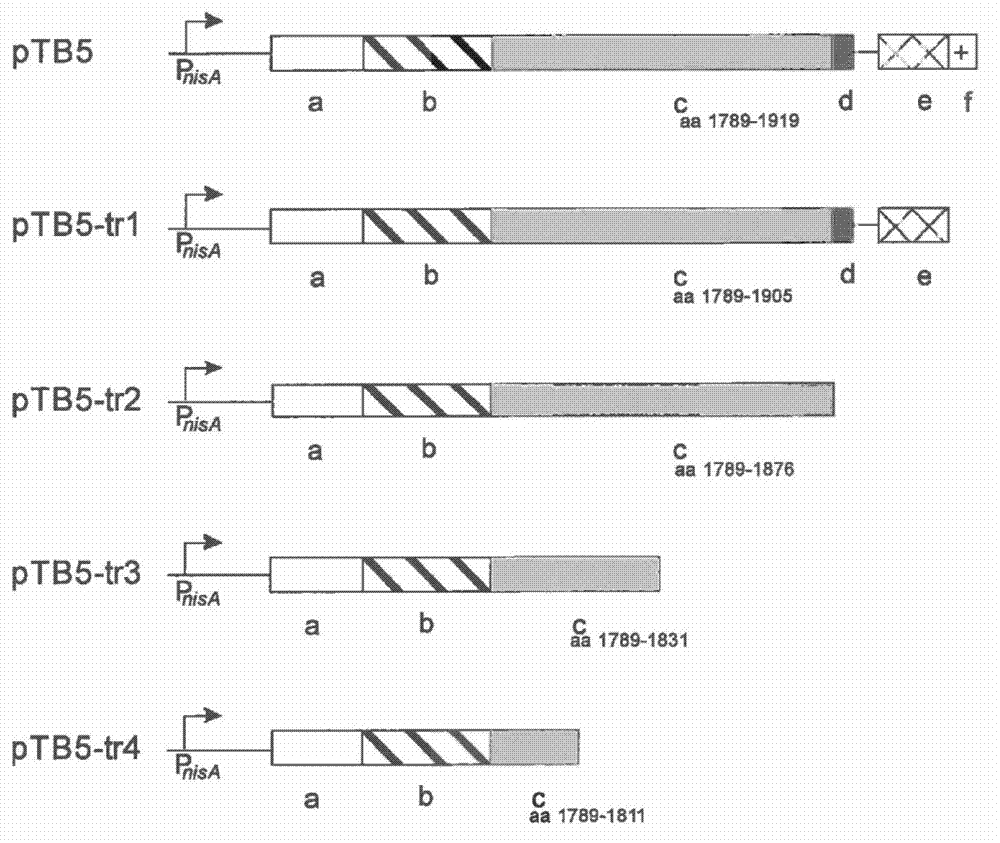
[0058] Example 1: Expression vectors for cell surface display of peptides containing thioether and dehydrogenation residues on Lactococcus lactis
[0059] Target : This example is about the construction of an expression vector for displaying a thioether-containing peptide on the surface of Lactococcus lactis. The Lactococcus lactis host organism provides the machinery for the introduction of thioether-linked nisin biosynthesis and export in the desired peptide and its export. The peptide is translationally fused to the LPXTG cell wall anchor motif, eg, Lactococcus lactis PrtP protease. This anchoring mechanism requires the processing of sortase for the covalent anchoring of peptides to the peptidoglycan of the bacterial cell wall. In this way, the peptide and the encoding DNA are linked, allowing selection / screening of post-translationally modified peptides with desired properties. Nisin will be used as a model peptide for the development and validation of the display syst...
Embodiment 2
[0063] Example 2: Antibodies against the leader peptide demonstrate attachment of the pronisin to the cell surface location of the cell wall of the fusion protein TB5.
[0064] Target : Evaluation of TB5 display on the cell surface of Lactococcus lactis by whole-cell ELISA using the anti-nisin leader peptide
[0065] Materials and methods
[0066] L. lactis NZ9000 (pTB5) and L. lactis NZ9000 (pIL3BTC / pTB5) cells were cultured in the presence or absence of nisin to induce TB5. After preparation, the produced cells were collected by centrifugation and washed three times with pH 7.4 phosphate-buffered saline (PBS). Equal numbers of TB5-displaying cells were incubated with 1000-fold diluted rabbit anti-nisin lead antibody solution in PBS containing 0.5% BSA in a final volume of 1 ml for 1 hr at room temperature with agitation. After three washes with PBS, the displayed TB5 was visualized by incubation with goat anti-rabbit IgG conjugated to alkaline phosphatase (1:10000) and...
Embodiment 3
[0069] Example 3: Prenisin targeting fusion proteins have been intracellularly modified with NisB and NisC prior to cell surface display. Modification by NisB- and NisC-modified prenisin-anchored fusion proteins was demonstrated by antibacterial activity against overlapping cells after leader peptide cleavage.
[0070] Target : Example 2 shows that production of TB5 results in the display of pro-nisin on the cell surface of Lactococcus lactis. In this example, nisin modification by NisB and NisC was assessed with an overlay of a nisin-sensitive Lactococcus lactis strain. Growth inhibition of this strain indicated that NisB and NisC correctly modified TB5 and formed at least loops A, B and C.
[0071] Materials and methods
[0072] Overlay of plaques with well-washed SDS-PAA gels or pTB5-producing L. lactis NZ9000 cells with 200-fold dilutions of L. lactis MG1363 or NZ9000 strains in 0.5% top agar containing 0.1 mg / ml trypsin GM17 agar plates. Trypsin is required for cl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com