Fluorescent staining kit for rapid detection on biological cell viability, and application of same
A biological cell, fluorescent dyeing technology, applied in measurement devices, particle and sedimentation analysis, individual particle analysis, etc., to achieve the effects of low toxicity, long exclusion culture period, and ease of use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] (1) Anthocyanin dyes are dissolved in dimethyl sulfoxide (DMSO) to a concentration of 5-6mM, which is the prepared concentrated solution of anthocyanin dyes for storage.
[0039](2) PI dye was dissolved in ultrapure water to 100 μg / mL, and stored away from light
[0040] (3) Pipette 1mL of Acetobacter xylinum solution into a 2mL centrifuge tube wrapped with tinfoil.
[0041] (4) Add 300 μL of anthocyanin dye diluted 1000 times with buffer and 200 μL of 100 μg / mL PI dye, shake up and down several times, and place in the dark for 10-30 minutes. The dilution buffer is 1×TE buffer (containing 10 mM Tris-HCl buffer in 1 mM EDTA, pH 8.0).
[0042] (5) Filter the stained bacterial solution through a 0.2 μm black filter membrane (Whatman), trap the bacteria on the black filter membrane, transfer the filter membrane to a glass slide, drop a drop of non-fluorescent essential oil in the center of the membrane, Cover with a coverslip.
[0043] (6) Observe the slide under the 100...
Embodiment 2
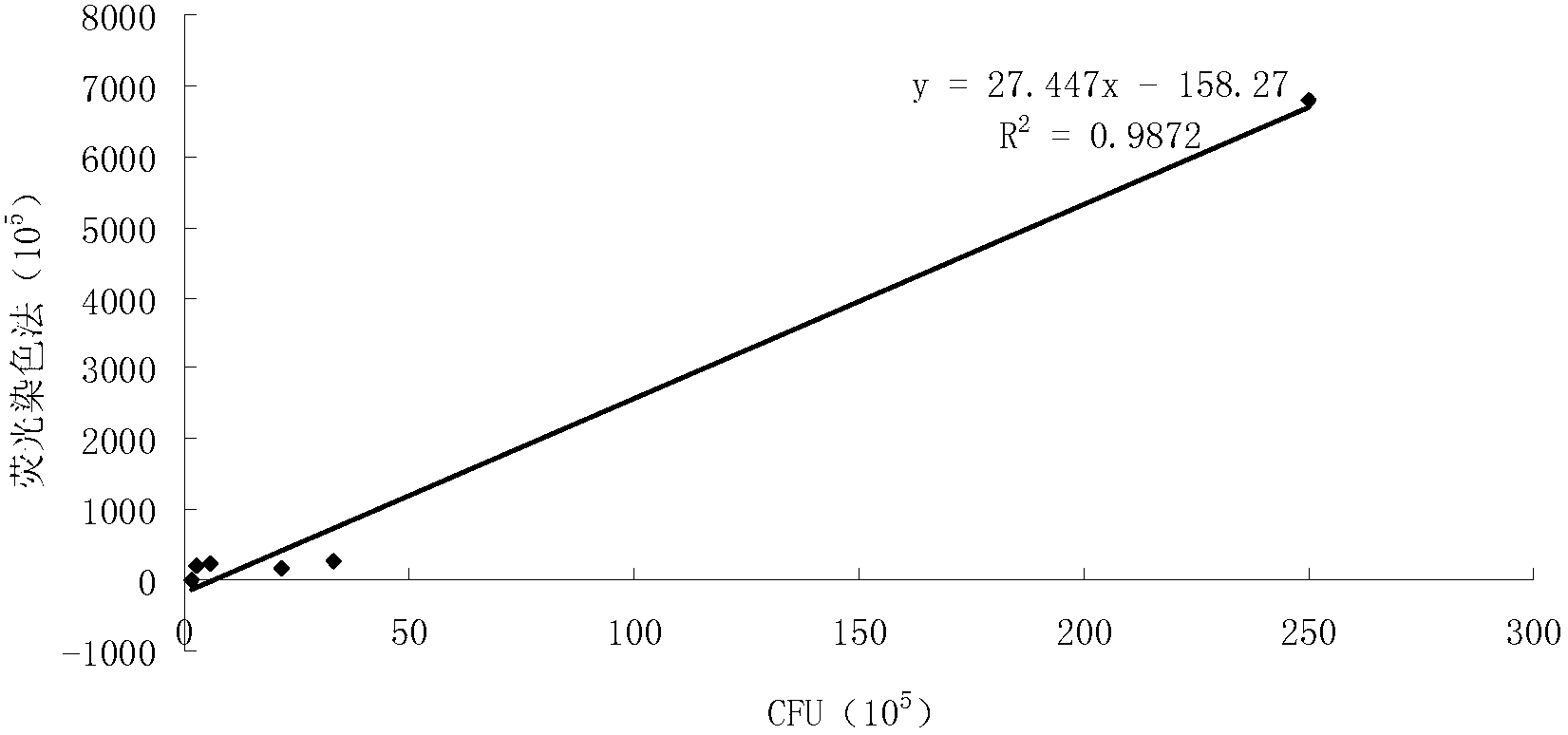
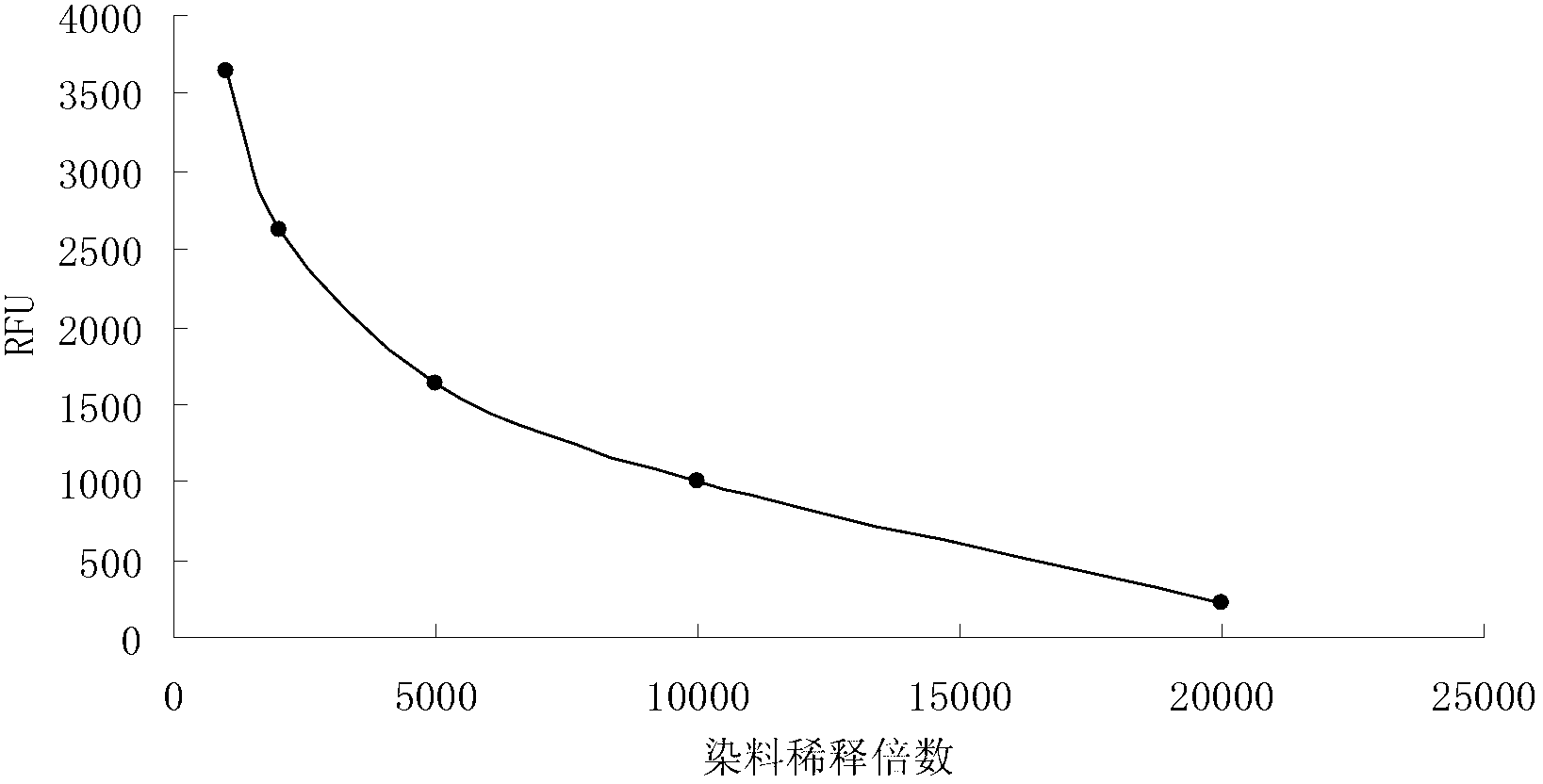
[0045] (1) Dilute the concentrated solution of anthocyanin dye storage by 1000 times to 20000 times, use dyes with the same volume but different dilution times to stain the bacteria according to steps 1-3 in Example 1, and use Molecular Devices Flexstation II calcium flow Fluorescence instrument was used to test, and it was found that the fluorescence intensity of the bacteria decreased with the increase of the dilution factor of the dye. When the dilution was 5000 times, the dyeing effect was better and the amount of dye was relatively low, which was more economical. If the dilution factor increases, the detected fluorescence value is close to the range of the error limit, and the error of the detection value increases (experimental results such as figure 2 ).
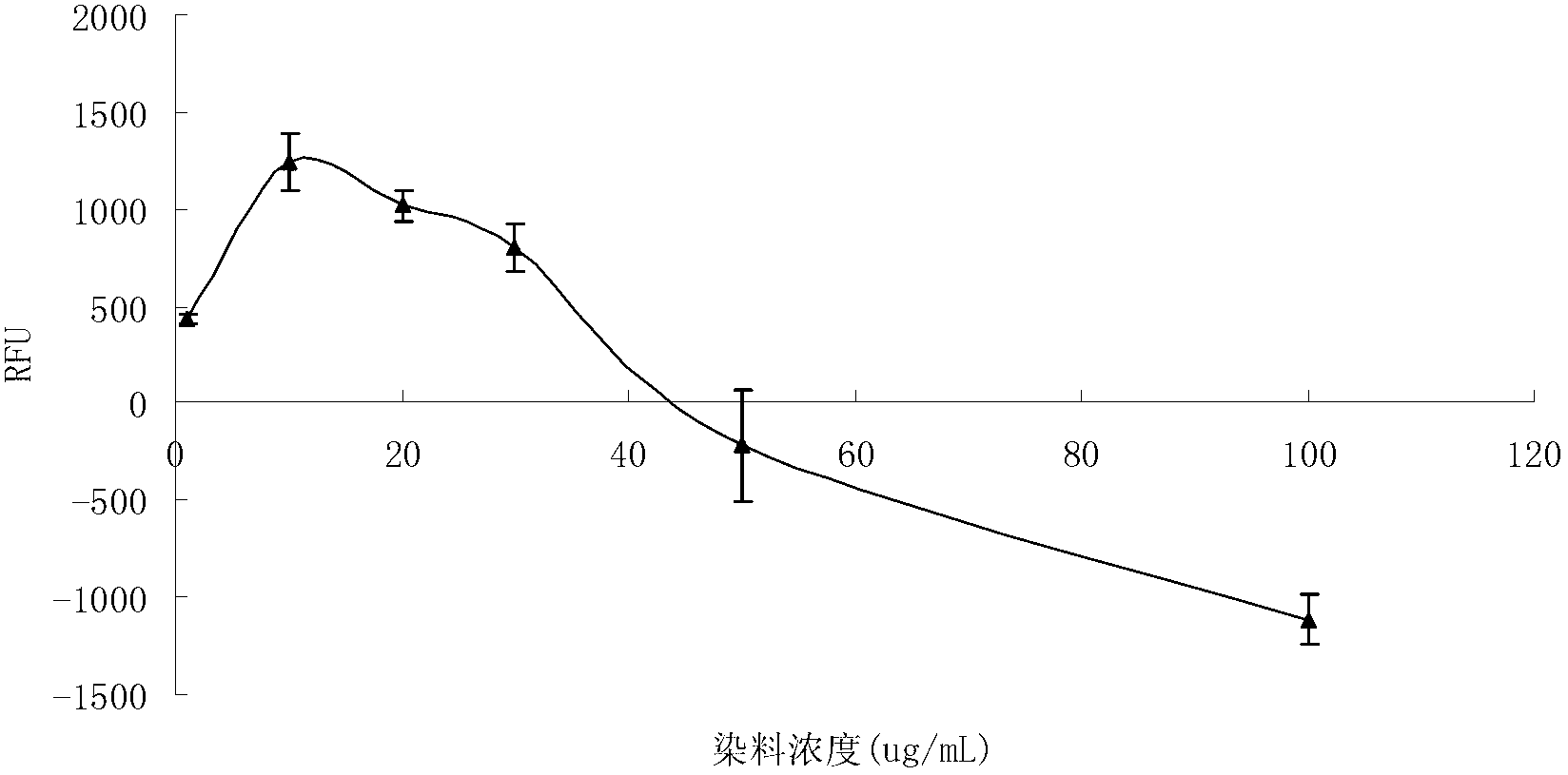
[0046] (2) The concentration of the dye PI was prepared from 1 μg / mL to 100 μg / mL, and the dyes with the same volume but different dilution times were used to stain the bacteria. It was found that the dyeing effect w...
Embodiment 3
[0048] (1) Cultivate Staphylococcus aureus until the turbidity OD value of the bacterial concentration is 0.7, take 25mL of the bacterial liquid and centrifuge at 12000r / min for 10-15 minutes;
[0049] (2) The collected bacteria were resuspended in 2 mL of normal saline;
[0050] (3) Dissolve 1mL of bacterial solution in 20mL of normal saline, and another 1mL in 20mL of 70% isopropanol;
[0051] (4) Stir every 15 minutes and incubate at room temperature for 1 hour;
[0052] (5) Centrifuge at 12000r / min for 10-15 minutes;
[0053] (6) Resuspend in 20mL of normal saline, then centrifuge and wash, the operation steps are the same as above;
[0054] (7) Resuspend with 10mL of normal saline respectively, and mix live bacteria and dead bacteria according to the ratio in Table 1;
[0055] Table I
[0056] Ratio of live bacteria to dead bacteria
[0057] (8) Add 140 μL of the mixed bacterial solution to the microwell plate, add 40 μL of 5 mM anthocyanin dye and 20 μL of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com