Method for detecting methylation level of synuclein intron 1
A synuclein and level detection technology, applied in the field of bioengineering, can solve the problems of decreased quantitative accuracy, time-consuming and laborious, and incapable of quantitative analysis, and achieve the effects of good quantitative characteristics, convenient operation, and high throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
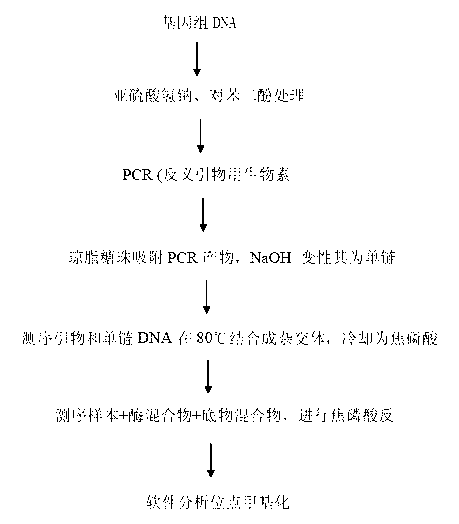
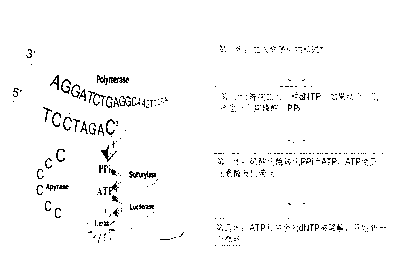
[0041] Example: Application of Pyrosequencing in α-Syn Intron 1 Methylation Detection
[0042] 1. Sodium bisulfite treatment
[0043] Genomic DNA is modified using sodium metabisulfite and hydroquinone to convert unmethylated cytosines to uracils, while methylated uracils remain unchanged. The process is briefly described as follows: first, the genomic DNA was denatured with 0.3mol / L NaOH; then, the genomic DNA was treated at 50°C for 16h with a mixed solution of sodium metabisulfite and hydroquinone (pH 5.0). The DNA was purified with a kit, and 0.3 mol / L NaOH was added to incubate at 37°C for 15 min to terminate the reaction, and finally the DNA was precipitated with ethanol.
[0044] 2. PCR amplification
[0045] Perform PCR amplification according to conventional methods, primers and reaction conditions are as follows:
[0046] Forward amplification primers: GAA, ATG, GAA, GTG, TAA, GGA, GGT, T;
[0047] Reverse amplification primers: Biotin-TTC, TAA, TCC, ATC, CAA, CA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com