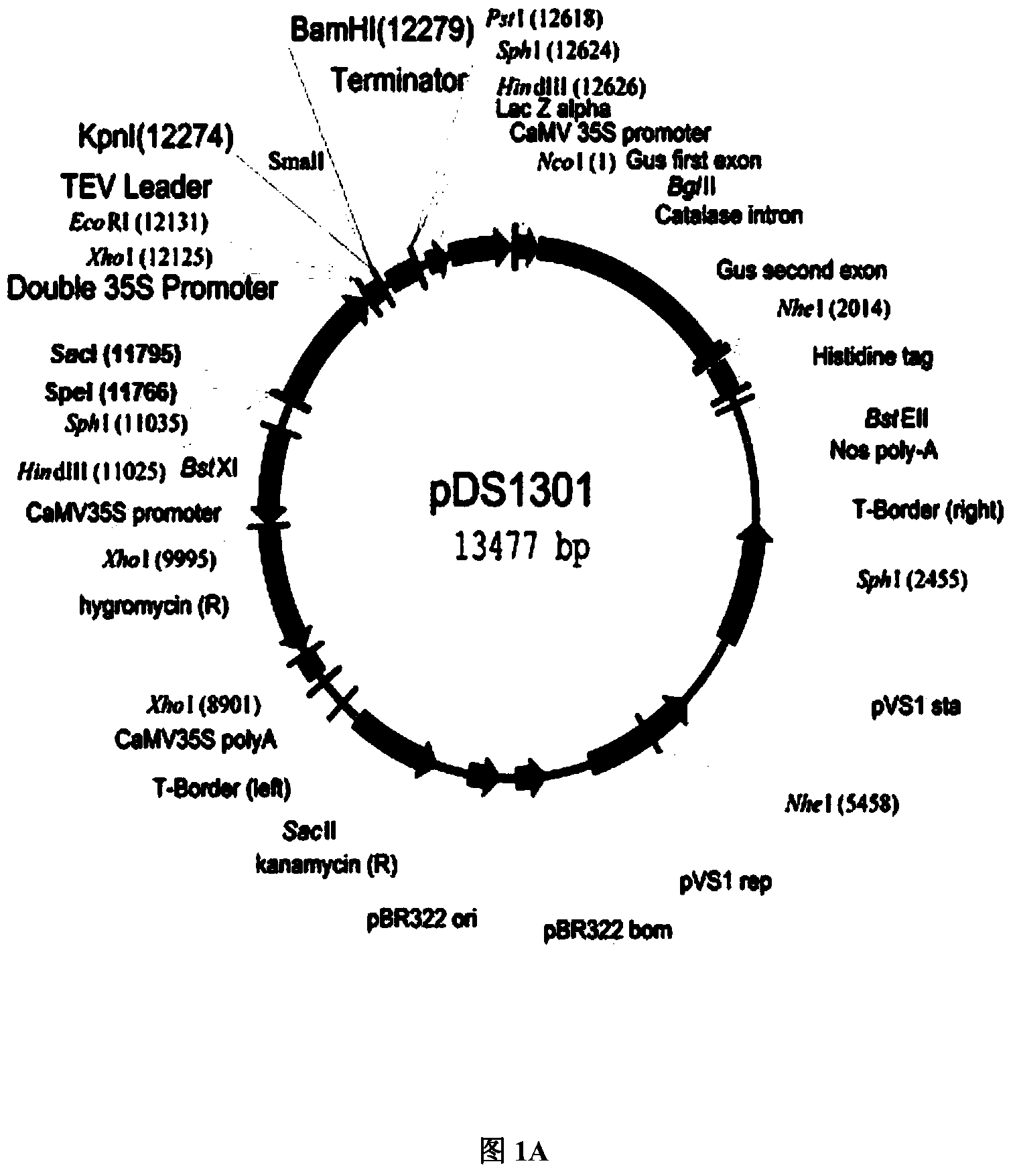
Application of histone deacetylase gene to regulation and control on development of rice seed starch
A deacetylase, rice seed technology, applied in application, genetic engineering, plant genetic improvement and other directions to achieve the effect of alleviating food shortages
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1: Amplification of the full-length cDNA of the OsSRT1 gene
[0036] For the gene OsSRT1 (gene accession number LOC_Os04g20270) required by the present invention, mainly by RT-PCR method (see: J. Experiment Guide (Third Edition), Beijing, Science Press, 2002 Edition) was amplified to obtain the full-length sequence of the OsSRT1 gene. The specific operation is as follows:
[0037] 1) Extract the RNA from the seedling leaves of the rice variety Huanghui 63. The reagent for RNA extraction is the Trizol extraction kit from Invitrogen (see the kit’s instruction manual for specific steps);
[0038] 2) The steps of reverse transcription to synthesize the first strand of cDNA in RT-PCR are as follows:
[0039] ① Prepare mixed solution 1: 4 μg of total RNA, 2U of DNaseI, 1 μl of 10×DNaseI buffer, add DEPC (diethyl pyrocarbonate, a strong inhibitor of RNase) treated water (0.01% DEPC) to 10 μl, mix well and mix Solution 1 was placed at 37°C for 20 minutes to remove DN...
Embodiment 2
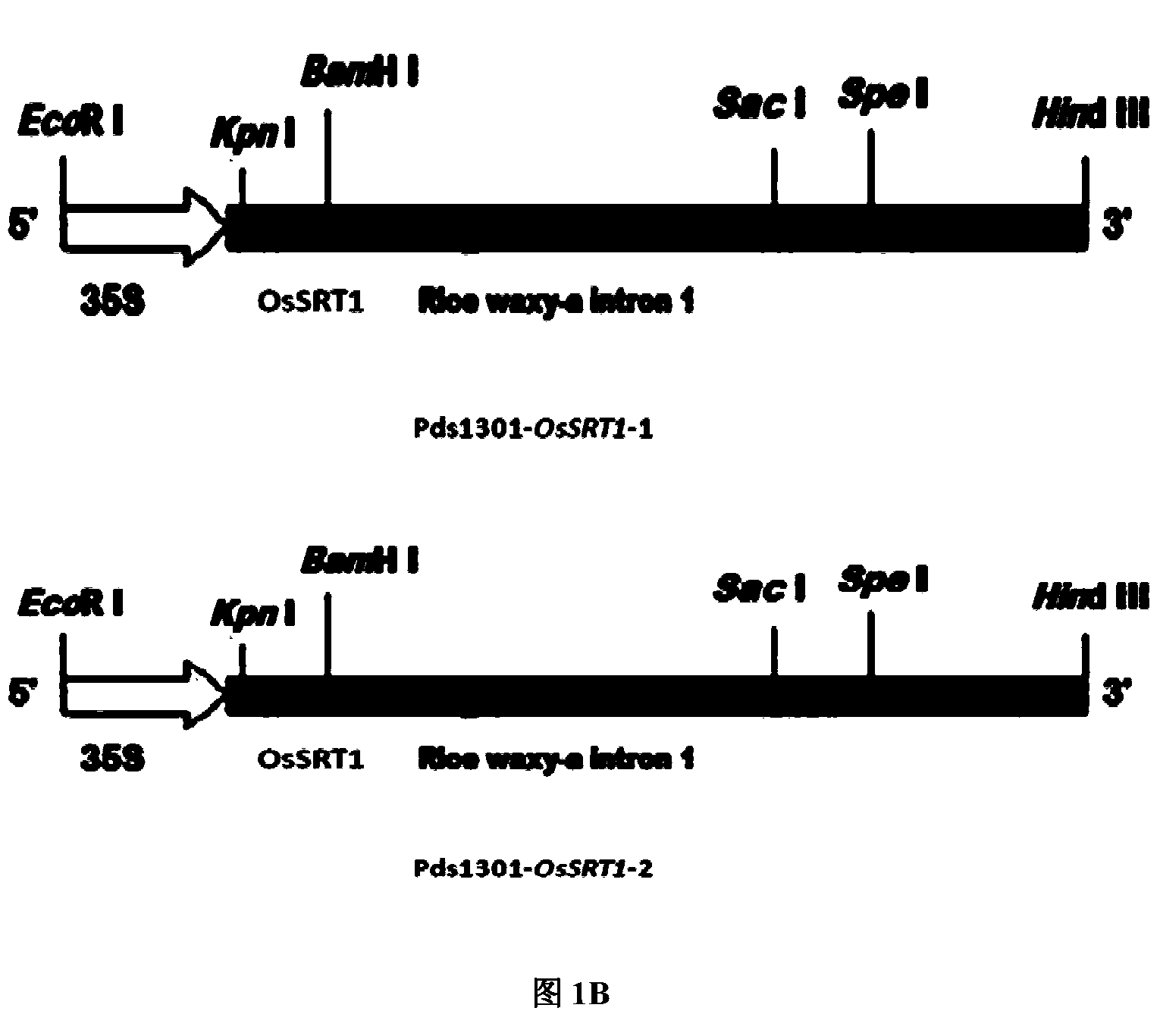
[0051] Example 2: Construction of OsSRT1 double-strand suppression vector
[0052] For the genes required by the present invention, by RT-PCR method (see: J. Sambrook, EF Fritsch, T Mani Artis, translated by Huang Peitang, Wang Jiaxi, etc., Molecular Cloning Experiment Guide (Third Edition), Science Press, 2002 edition) to amplify to obtain a specific sequence of OsSRT1. The specific steps are: by using the full-length cDNA of OsSRT1 obtained by self-amplification as a template, find the specific sequence of OsSRT1 to design primers, and PCR amplifies the RNAi inhibitory fragment. The amplified product was connected to pGEMT-vector (purchased from Promega (Beijing) Biotechnology Co., Ltd., ie Promega, USA) by T / A cloning for sequencing verification.
[0053] The primers used to clone the RNAi suppressor fragment of OsSRT1 are as follows:
[0054] ds-F:5'-GGGACTAGTGGTACCAGTCCTGCAAGAGTTGCAAC-3'
[0055] ds-R:5'-GGGGAGCTCGGATCCCCAGCTTTCACATGCACTAG-3'
[0056] Specific steps ar...
Embodiment 3
[0061] Example 3: Transformation of binary Ti plasmid vector and detection of positive and expression levels of transgenic plants
[0062] 1) The newly constructed suppression expression vector pDS1301-OsSRT1-2 (from Example 2) was introduced into Agrobacterium EHA105 (purchased Among the strains from CAMBIA Laboratory in Australia, the strain transformed with pDS1301-OsSRT1-2 was named pDS1301-OsSRT1-2-EHA105.
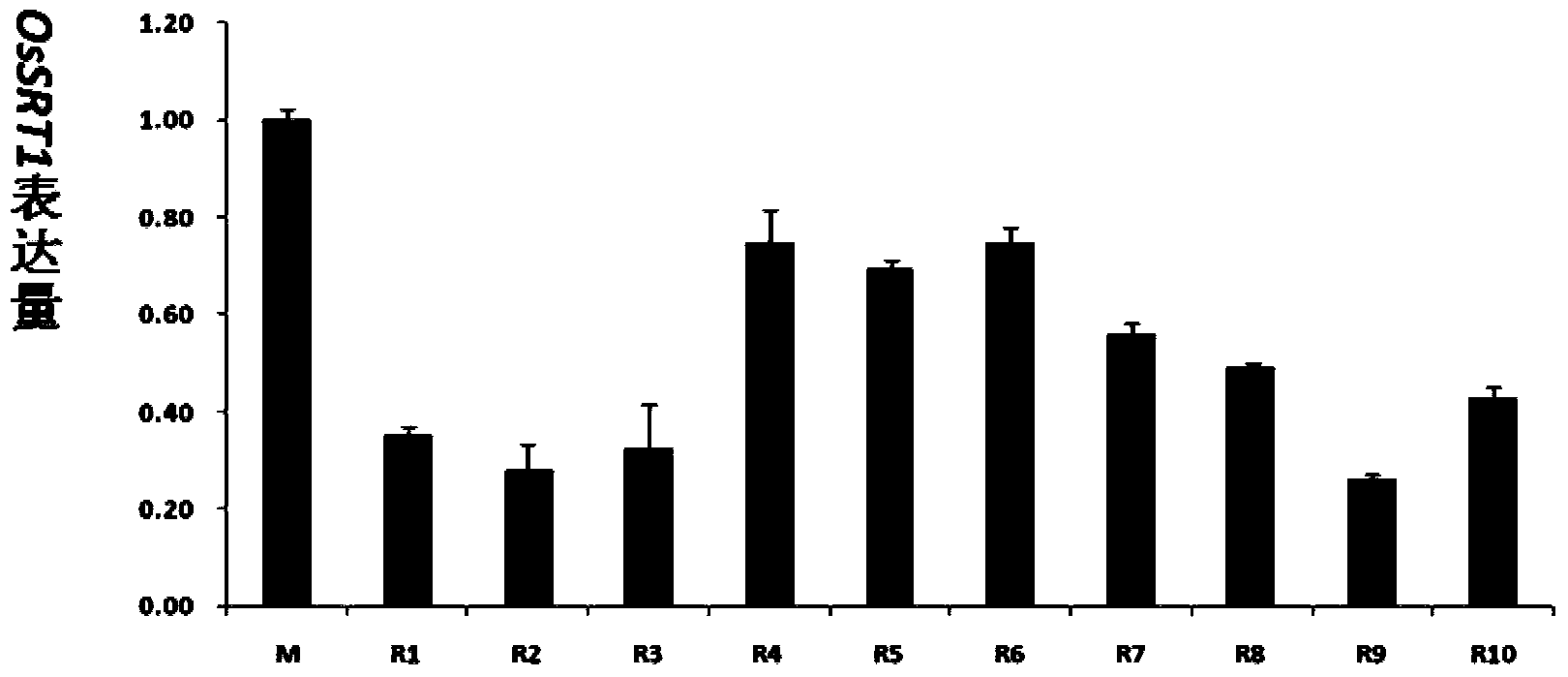
[0063] 2) Transform the pDS1301-OsSRT1-2-EHA105 obtained in the previous step into the rice variety Minghui 63. The transformation method refers to the method reported by Hiei et al. (Hiei et al., Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J, 1994, 6:271-282.). The obtained T0 transgenic plants were named Rn, where n=1, 2, 3...represent different transgenic families.
[0064] 3) Total DNA was extracted from the leaves of the transformed plants of the T0 generation. The DNA ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com