Porcine Sapelovirus real-time fluorescent quantitative RT-PCR detection method
A real-time fluorescence quantitative and detection method technology, applied in the biological field, can solve problems such as inability to detect, time-consuming, and delay in disease control
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
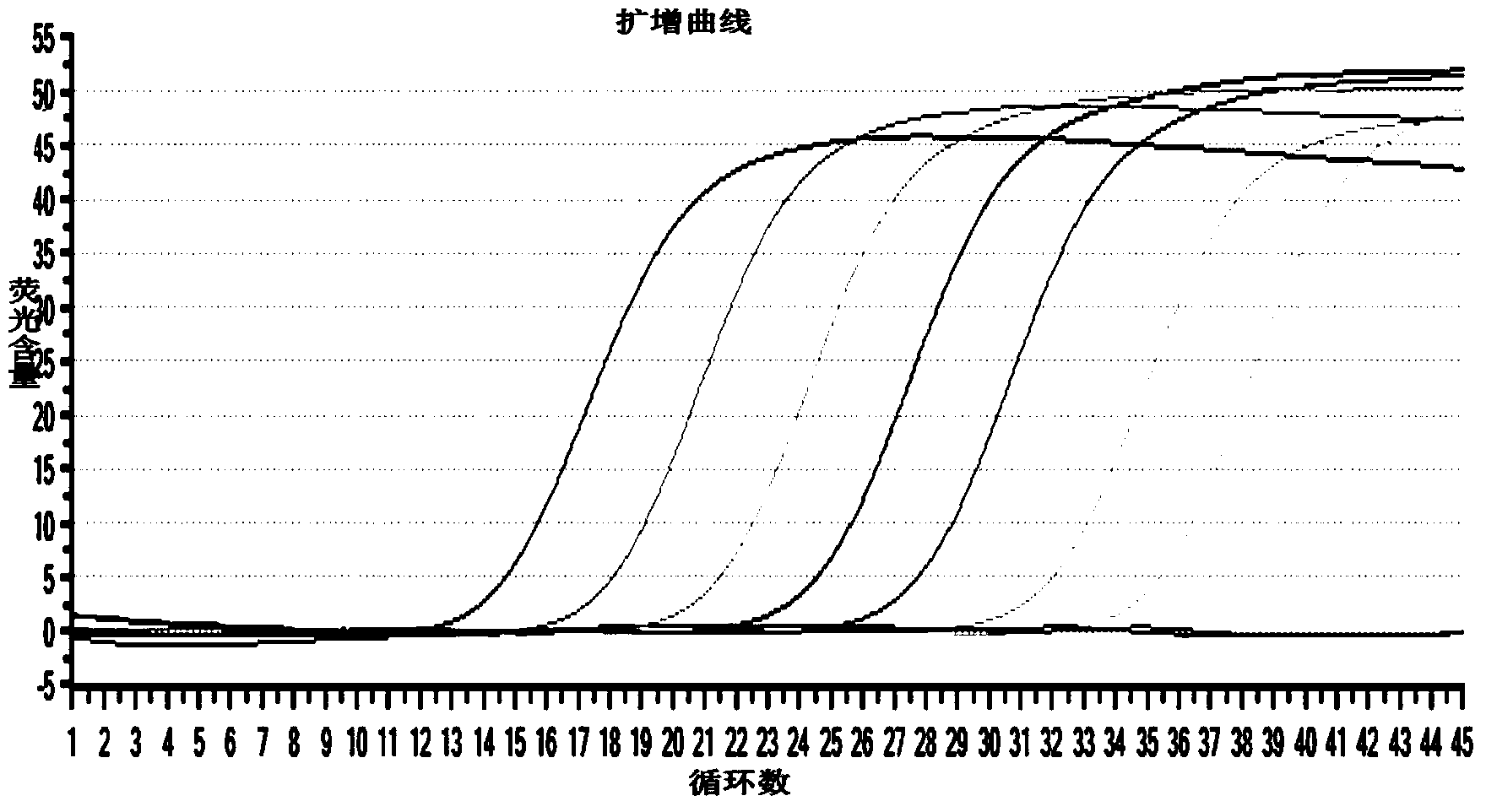
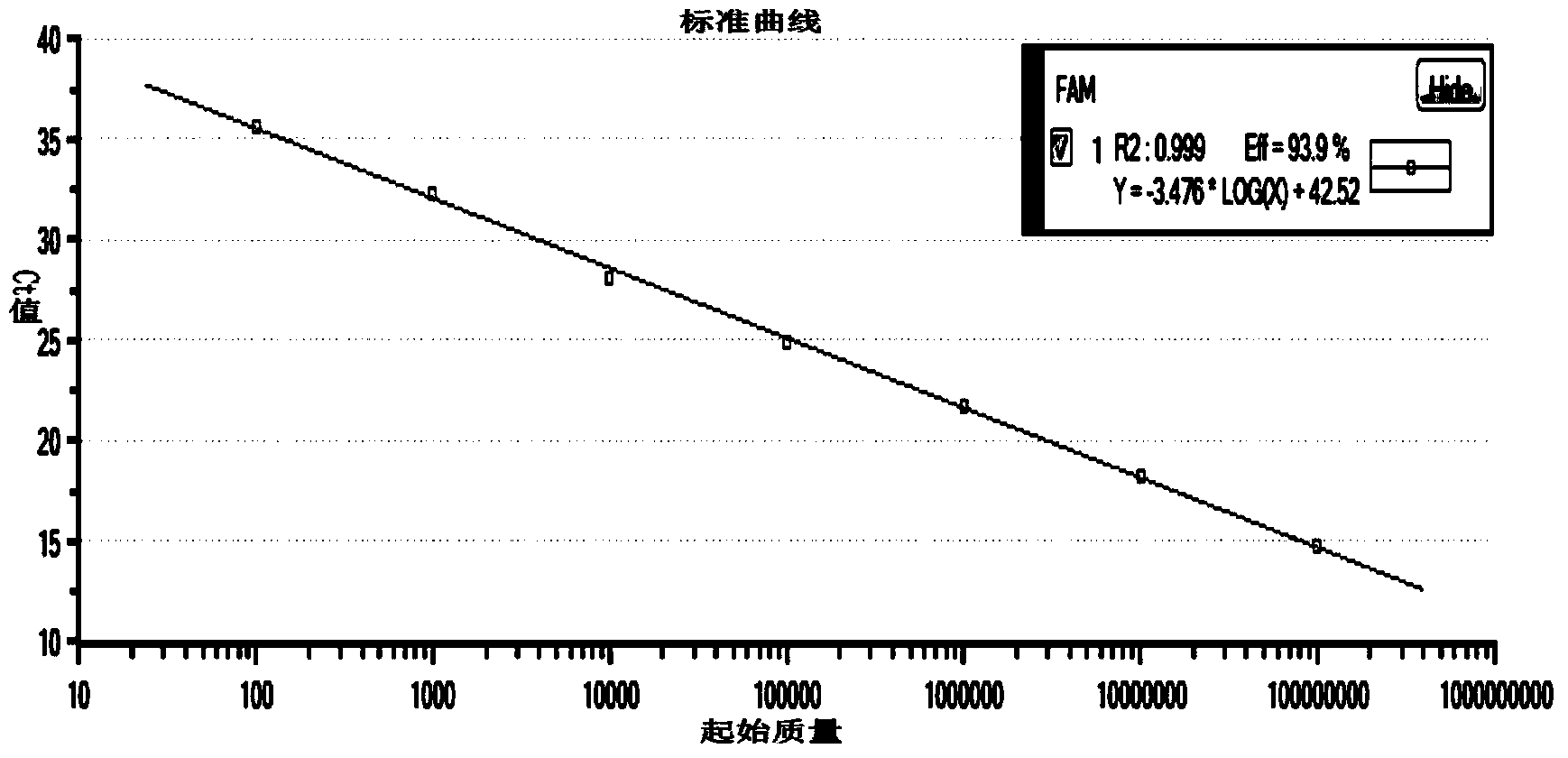
[0050] The making of embodiment 1 standard curve
[0051] 1) Primer design for the construction of amplified fragment standards
[0052] Design and construct primers on the target gene, and add a T7 promoter to the 5' end of the constructed upstream primer, and add 25 Ts to the 3' end of the constructed downstream primer, so that the constructed target gene standard RNA has Poly(A )tail. The T7 promoter sequence is as follows: ACTAATACGACTCACTATAGGG
[0053] The positional relationship of the specific primer design is shown in the following sequence (the spaced part is the construction primer region, the underlined part is the fluorescent quantitative RT-PCR primer region, the wavy part is the T7 promoter and the Poly(A) tail, and the arrow indicates the transcription start position ).
[0054] TCCCTTCCAAAGC GGATGGACACAAGGACTTGGACTCACGGCAAGTGCACATGGTATGATTTTGGATACACCTTAACGGCAGTAGCGTGGCGAGCTATGGAAAAATCGCAATTGTCGATAGCCATGTTAGTGACGCGCTTCGGCGTGCTCCTTTGGTGATTCGGCGACTGGTTACA ...
Embodiment 2
[0072] Embodiment 2 specificity experiment
[0073] Select nucleic acid standards of porcine Jieshen virus, porcine enterovirus 9, porcine foot-and-mouth disease virus, encephalomyocarditis virus, porcine transmissible gastroenteritis, porcine epidemic diarrhea, porcine reproductive and respiratory syndrome virus that do not contain Sapero virus , configure the reaction solution according to the above method, set the reaction conditions, and perform the reaction in Applied Biosystems7500 / 7500 Fast Real-Time PCR system. According to the amplification curve, only porcine Sapero virus was amplified, indicating that the primer had good specificity.
Embodiment 3
[0074] Embodiment 3 clinical verification
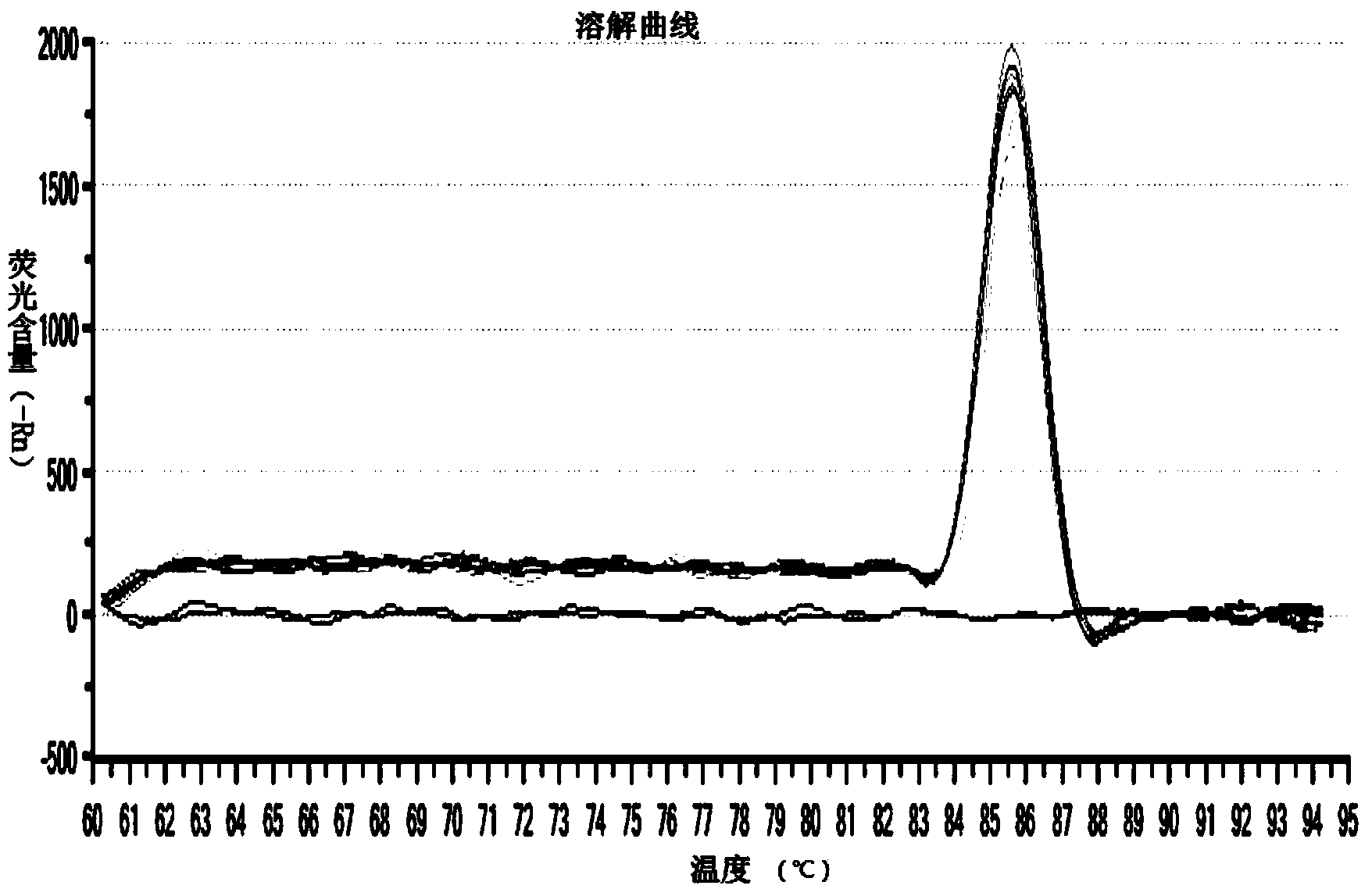
[0075] We collected 63 fecal samples from the Zhengyi Pig Farm in Minhang District, Shanghai and the Qibao Campus Pig Farm of Shanghai Jiaotong University. After treatment with 1% phosphate buffer, RNA was extracted using a commercial RNA / DNA extraction kit (Tiangen Biochemical Technology (Beijing) Co., Ltd.). Configure the reaction solution according to the above method, set the reaction conditions, and perform the reaction in Applied Biosystems7500 / 7500 Fast Real-Time PCR system. The results showed that 7 samples were amplified, and the Ct value was less than 35, and the Tm value was within the range of 85.62±0.11, which were considered positive samples, with a positive rate of 11.11%. The same samples were verified by ordinary RT-PCR. RT-PCR results showed that amplification occurred in 5 samples, and the positive rate was 7.93%. It shows that SYBR real-time fluorescent quantitative RT-PCR is more sensitive than RT-PCR method, a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com