Industrial rice wine yeast metabolic engineering bacteria with low-yield urea and building method thereof
A rice wine yeast and metabolic engineering technology, applied in fungi, introduction of foreign genetic material using vectors, recombinant DNA technology, etc., can solve the problems affecting the growth and fermentation ability of Saccharomyces cerevisiae, low transformation efficiency and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1 Construction of uracil-deficient yeast engineering strain Δura3
[0045] 1. Construction of URA3 gene knockout module
[0046] Extract the genomic DNA of the parental strain (see the instruction manual of the MiniBEST Universal Genomic DNA Extraction Kit kit for specific operation methods); use the genomic DNA as a template, and use Prime STAR DNA Polymerase to amplify the upstream of the URA3 gene using primer pairs P1 / P2 and P3 / P4, respectively The homology arm (URA3L) and the downstream homology arm (URA3R), and then use P1 / P4 primers to fuse URA3L and URA3R by fusion PCR to obtain the URA3 gene knockout module (URA3L-URA3R) (the specific operation method and reaction system of fusion PCR Please refer to Prime STAR DNA Polymerase instructions for preparation).
[0047] The amplification procedure for each primer pair is as follows:
[0048] (1) P1 / P2 and P3 / P4: Pre-denaturation at 95°C for 4 minutes, (denaturation at 98°C for 10 s, annealing at 52°C for 1...
Embodiment 2
[0064] Example 2 Construction of diploid industrial yellow rice wine yeast engineering bacteria with high expression of urea amidase gene (DUR1, 2)
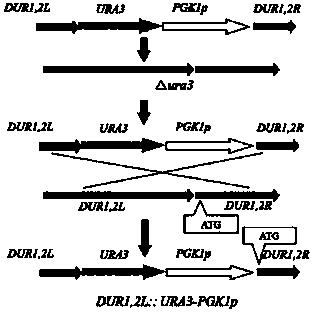
[0065] 1. Construction of high expression module DUR1,2L-URA3-PGK1p-DUR1,2R containing PGK1p strong promoter and URA3 gene
[0066] For a schematic diagram of the build, see image 3 . First, using the genomic DNA of industrial rice wine yeast as a template, Prime STAR DNA Polymerase was used to amplify the URA3 gene, PGK1p strong promoter, DUR1,2 initiation Homology arm before the codon (DUR1,2L), homology arm after the start codon of DUR1,2 (DUR1,2R). Then, using the equimolar four fragments as a template, the above four fragments were fused by landing PCR without primers. The PCR reaction system is shown in Table 2:
[0067] Table 225μL PCR reaction system
[0068]
[0069] The PCR amplification program is as follows: pre-denaturation at 95°C for 4 min, then enter the PCR amplification program, denaturation at 98°C for...
Embodiment 3
[0084] Embodiment 3 yellow rice wine fermentation experiment
[0085] The diploid yeast engineered bacterium obtained in Example 2 is carried out rice wine fermentation experiment together with parent bacterial strain to investigate the ability of engineered bacterium to reduce urea and EC content, concrete operation steps are as follows:
[0086] (1) Soak rice
[0087] Take 50g of glutinous rice, add water to soak the rice layer for about 10cm, and soak for 48 hours at room temperature;
[0088] (2) Cooking
[0089] Cook the soaked rice under normal pressure, and keep it for 25 minutes after a large amount of steam comes out, until the grains are uniform, there is no white heart inside, cooked but not sticky, soft but not rotten;
[0090] (3) Bibimbap
[0091] Cool the rice in a ventilated place, spread it to about 27°C, add 1.5g of cooked wheat koji, 7g of raw wheat koji, and 85g of water;
[0092] (4) Pre-fermentation
[0093] The fermented mash was added to the activa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com