Primer pair and probe used for detecting phytophthora hibernalis and detection method for phytophthora hibernalis
A technology of Phytophthora solani and primer pair, which is applied in the field of molecular biology detection to achieve the effects of good specificity, simple operation and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] The design and synthesis of embodiment 1 primer pair and probe
[0024] According to the ITS sequence of Phytophthora winteris, the primer pair and probe were designed using the probe design software Express 3.0. The sequence of the primer pair is:
[0025] PH-F (forward primer): 5'-CGACTTGCCACCGGGA-3'
[0026] PH-R (reverse primer): 5'-AACGGTACTTCTCTTTGCTCGAA-3'
[0027] The sequence of the probe is: 5'-F1-TTCCACAACCAATTCCAT-Q1-3'; wherein, F1 is a FAM fluorescent dye, and Q1 is a non-fluorescent quencher group MGB.
[0028] The above primer pairs and probe sequences were synthesized by Shanghai Jikang Biotechnology Co., Ltd.
Embodiment 2
[0029] The extraction of embodiment 2 sample total DNA
[0030] Include the following steps:
[0031] 1) Collect an appropriate amount of Phytophthora hibernalis fungal mycelium or infected fruits and leaves, grind it into powder with liquid nitrogen, take 0.4g and put it into a 1.5mL centrifuge tube.
[0032] 2) Preheat the CTAB extract to 65°C, add 600 μL of the preheated CTAB extract to the 1.5 mL centrifuge tube containing the powder, and mix well.
[0033] 3) Immediately put in a water bath at 65°C for 30 minutes, then add 500 μL of chloroform / isoamyl alcohol (24:1, volume ratio), turn it upside down several times, and centrifuge at 13,000 g for 1 minute.
[0034] 4) Pipette about 400 μL of the supernatant into a new 2.0ml centrifuge tube, add 500 μl of chloroform / isoamyl alcohol (24:1), 500 μL of CTAB extract, mix gently, and centrifuge at 13000g for 1 minute.
[0035] 5) Take 800μl supernatant to a new 1.5mL centrifuge tube, add 500μL isopropanol, mix gently, and plac...
Embodiment 3
[0038] Example 3 Establishment of real-time fluorescence quantitative PCR (fluorescence RT-PCR) method
[0039] 1. Fluorescence RT-PCR reaction system
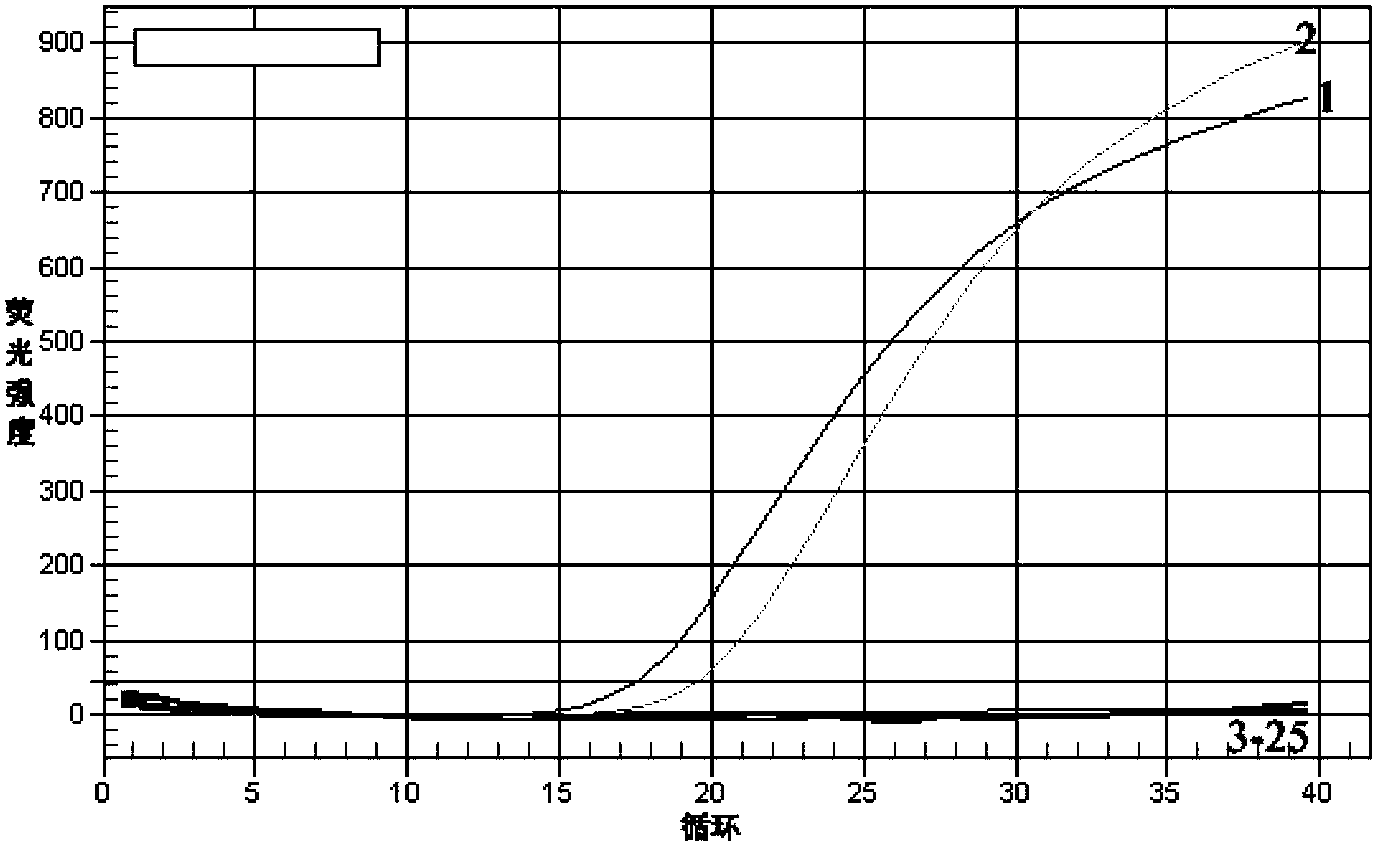
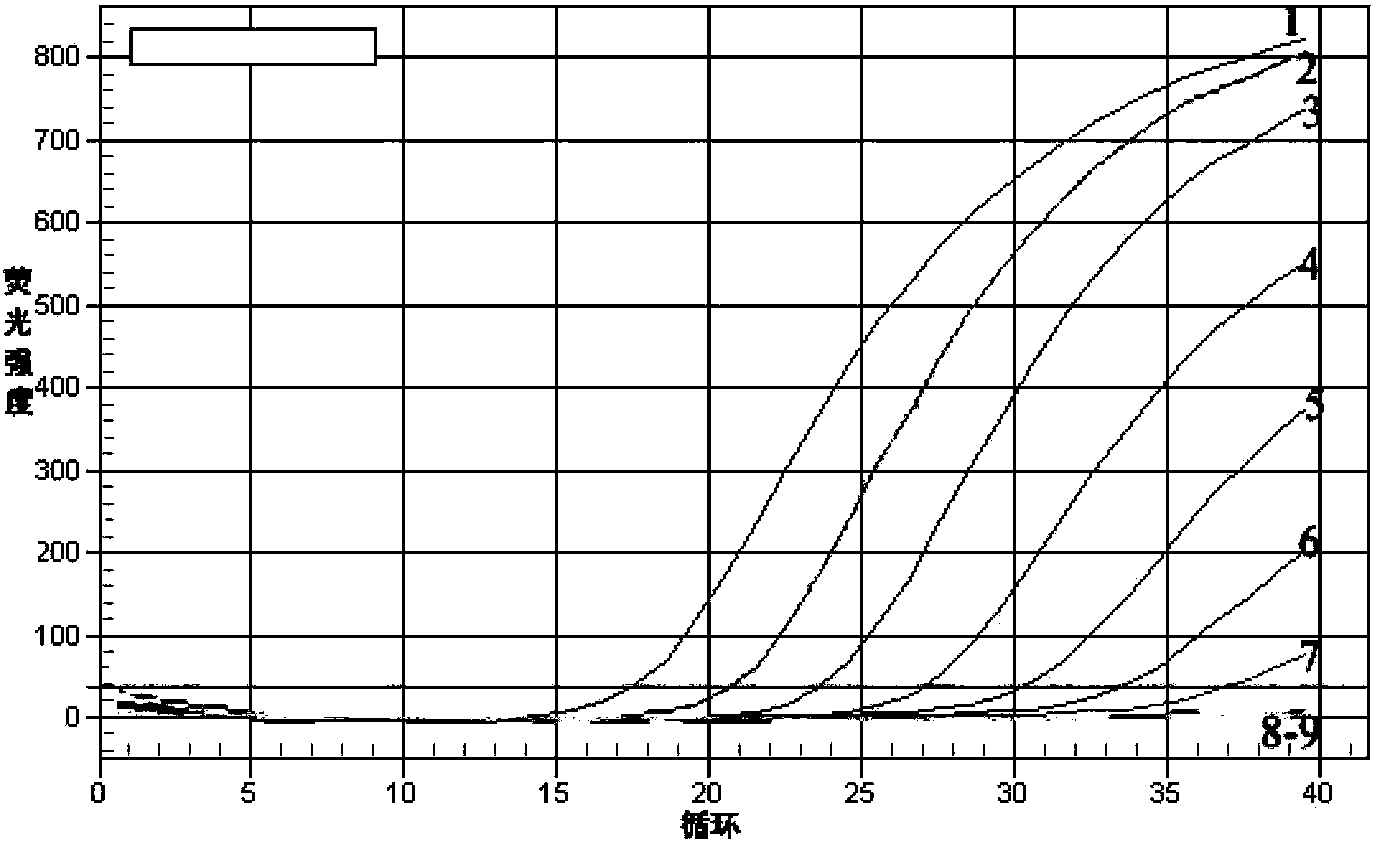
[0040] Using the total DNA as a template, carry out real-time fluorescent PCR reaction, the reaction system (total volume 25μL) is: sample DNA 1μL (0.0005-50ng), 10×PCR reaction buffer (without Mg 2+ ) 2.5 μL, 25 mM MgCl 22.0 μL, 2.5 mM dNTP 2.0 μL, 10 μM primer PH-F 1 μL, 10 μM primer PH-R 1 μL, 10 μM probe 1.5 μL, 5U / μL Taq DNA polymerase 0.3 μL, ddH 2 O 13.7 μL.
[0041] 2. Real-time fluorescent PCR reaction conditions
[0042] Place the sample tube into the BIO-RAD IQ TM 5. After the fluorescent PCR instrument, set the following conditions for the reaction: the first cycle is 95°C for 3min; then 95°C for 10s, 60°C for 30s, 40 cycles. Collect data after each cycle. After the reaction, judge the result according to the amplification curve.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com