Deoxyribonucleic acid (DNA)-modified gold nanoparticle colorimetric sensor and preparation method and use thereof
A colorimetric sensor and gold nanotechnology, applied in the field of bioanalysis, achieves high selectivity, low energy consumption, and simple preparation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
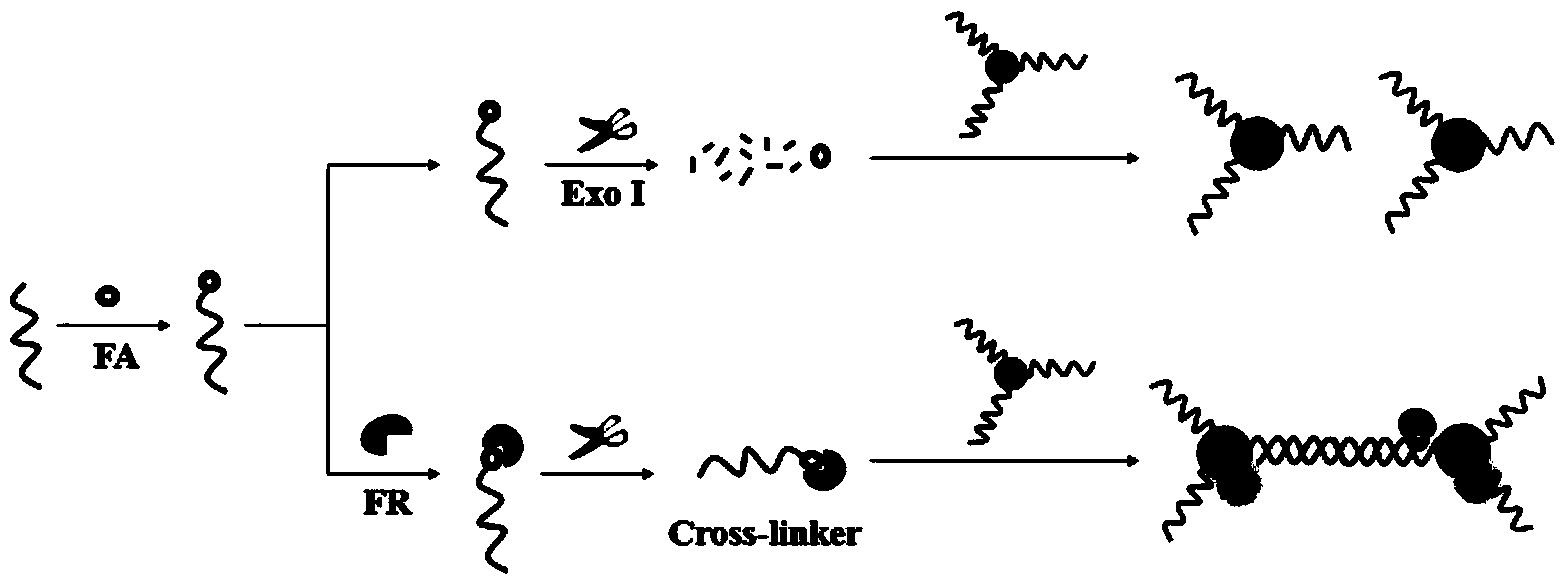
[0035] A DNA-modified gold nanocolorimetric sensor for the colorimetric detection of folate receptors based on tail protection. A method for preparing a gold nanoparticle colorimetric sensor, specifically comprising:
[0036] a. Dissolve the purchased DNA sequences (thiol: S1, FA-DNA: S2) in 0.05M Tris-HCl (pH7.4) buffer solution, and store them at 4°C for future use.
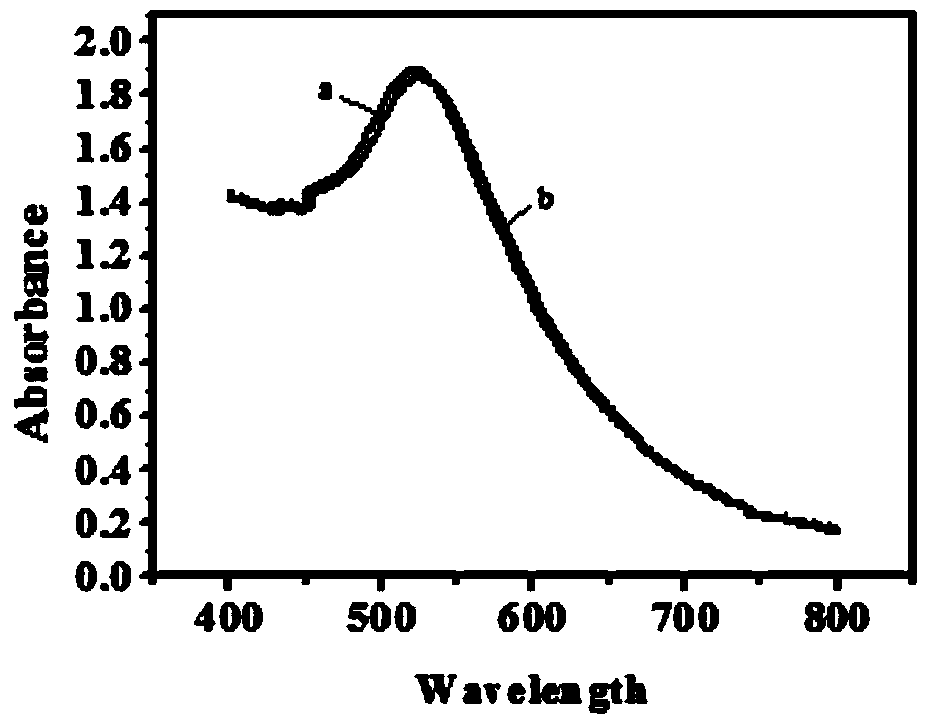
[0037] b. Using the succinimide coupling method (EDC-NHS) to bind FA to S2 with a modified amino group at the 3' end. 0.5mL of 20μM S2 was mixed with 0.5mL of 100mM Tris-HCl buffer solution (pH7.4, containing 10mM FA, 1mM EDC, 5mM Sulfo-NHS) and incubated at 37°C in the dark for 3h. The FA-S2 solution was dialyzed in PBS buffer solution (pH 7.4) to remove excess FA (molecular weight cutoff: 1000 Da). Then FR with standard serial concentration was added to the above-mentioned dialyzed solution, and cultured at 37° C. in the dark for another 2 h. Finally, 400U / mL Exo I was added to the above mixed solution, an...
Embodiment 2
[0040] A DNA-modified gold nanocolorimetric sensor for the colorimetric detection of folate receptors based on tail protection. The preparation and application steps of the colorimetric sensor based on S1-AuNP and S2 / FA-FR are as follows:
[0041] a. Dissolve the purchased DNA sequences (thiol: S1, FA-DNA: S2) in Tris-HCl (pH7.4) buffer solution, and store them at 4°C for future use.
[0042] b. Using the succinimide coupling method (EDC-NHS) to bind FA to S2 with a modified amino group at the 3' end. A certain concentration of S2 and Tris-HCl buffer solution (pH7.4, containing 10mM FA, 1mM EDC, 5mM Sulfo-NHS) were mixed evenly in a certain ratio, and incubated in the dark at 37°C for several hours. The FA-S2 solution was dialyzed against PBS buffer solution (pH 7.4) to separate excess FA (molecular weight cutoff: 1000 Da). Then add standard serial concentration of FR into the above dialyzed solution, and incubate for a period of time in the dark at 37°C. Finally, 400U / mL E...
Embodiment 3
[0045] Preparation of S1-Au NPs: Using a chemical reduction method, first synthesize a dispersion solution of Au NPs with uniform size (14nm), and then modify DNA-S1 with sulfhydryl groups to the surface of the particles.
[0046] The preparation and application steps of the colorimetric sensor based on S1-Au NPs and S2 / FA-FR are as follows:
[0047] a. Dissolve the purchased DNA sequences (thiol: S1, FA-DNA: S2) in 0.05M Tris-HCl (pH7.4) buffer solution, and store them at 4°C for future use.
[0048]b. Using the succinimide coupling method (EDC-NHS) to bind FA to S2 with a modified amino group at the 3' end. 0.5mL of 20μM S2 was mixed with 0.5mL of 100mM Tris-HCl buffer solution (pH7.4, containing 10mM FA, 1mM EDC, 5mM Sulfo-NHS) and incubated at 37°C in the dark for 3 hours. The FA-S2 solution was dialyzed against PBS buffer solution (pH 7.4) to separate excess FA (molecular weight cutoff: 1000 Da). Then FR with standard serial concentration was added to the above-mentione...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com