Molecular markers for Xanthomonas oryzae and application thereof
A Xanthomonas and molecular marker technology, applied in the application field of detecting two kinds of pathogenic bacteria, can solve the problems of poor identification accuracy and unreliable identification technology, and achieve the effect of improving detection reliability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1 Design of molecular markers that can distinguish and identify rice bacterial blight and rice bacterial streak
[0023] Genomes of three races KACC10331, PXO99A, MAFF311018 of Xanthomonas oryzae pv. oryzae and two races BLS256 and RS105 of Xanthomonas oryzae pv. oryzicola Sequences and related information were downloaded from NCBI (http: / / www.ncbi.nlm.nih.gov / ). (Table 1)
[0024] Table 1 Statistics of five bacterial genome information
[0025]
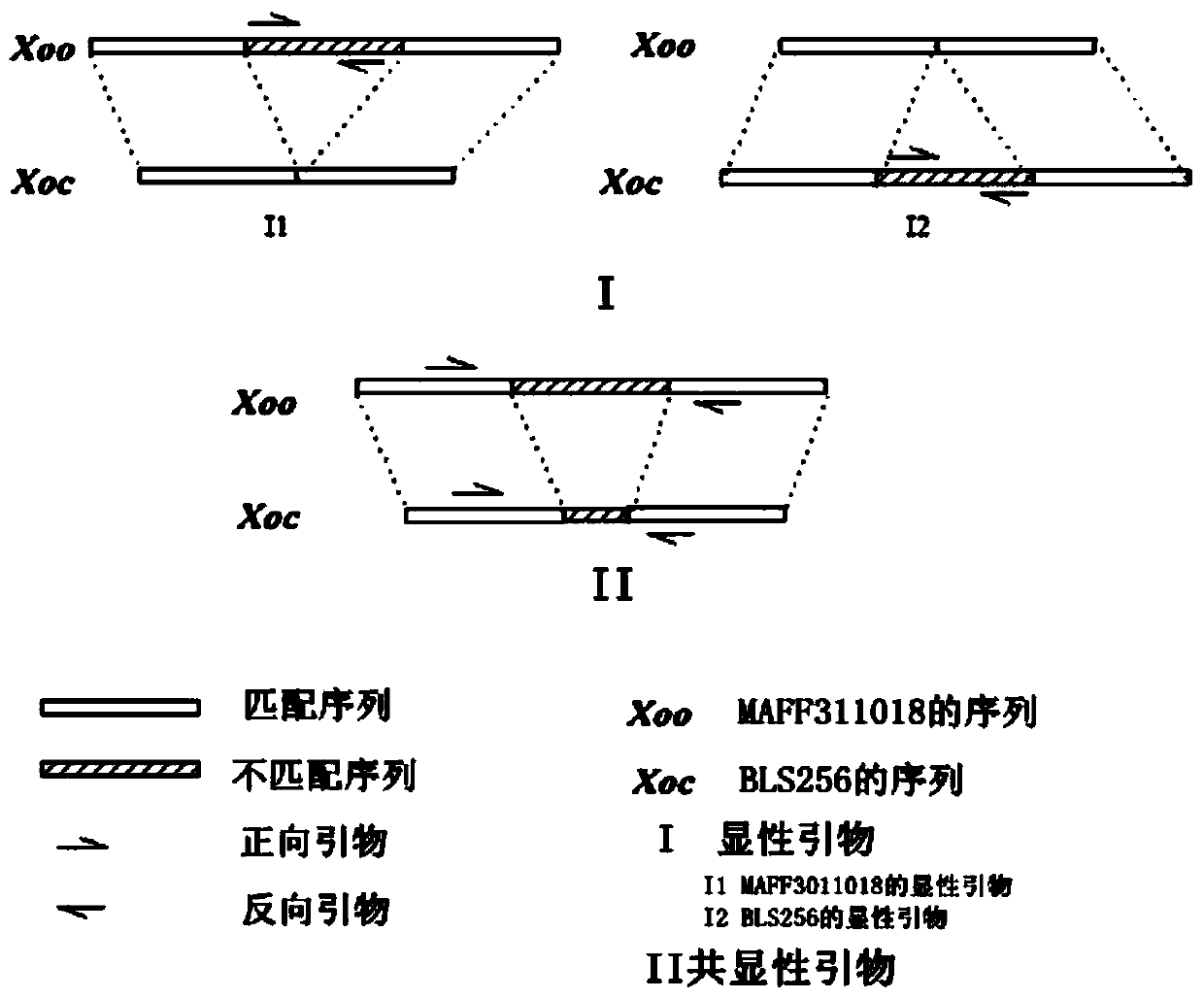
[0026] The inventors compared the whole genomes of three races of Xoo (MAFF311018, KACC10311, PXO99A) and found that the sequence similarity was high, so MAFF311018 was selected as the model race of Xoo.
[0027] Compared with RS105 and BLS256, it was found that BLS256 contained all the information of RS105, with a high degree of similarity, so BLS256 was used as the model race of S. oryzae.
[0028]BLAST was used to compare the whole genome sequences of the two model races MAFF311018 and BLS256, and the perl pro...
Embodiment 2
[0038] Example 2 Screening and verification of dominant molecular markers for rice bacterial blight specialization
[0039] According to the ePCR screening results of the designed specific primers for Xanthomonas oryzae, we selected 40 pairs of primers to verify against 25 strains of Xanthomonas oryzae and 10 strains of Bacterial blight of rice and other related strains .
[0040] Streak the strains to be tested on the TSA solid medium plate, culture at 28°C for 2 days, pick a single colony, place it in TSA liquid medium, and heat it at 180r min -1 , 28 ° C shaker culture for two days. Aspirate 3mL of the bacterial solution, and use the kit to extract the genomic DNA of the strain. And use it as a template for PCR reaction.
[0041] The optimized reaction system is: Taq enzyme 0.2 μL (5u·μL -1 ), 10×PCR Buffer (Mg 2+ Free) 2μL, 25mM MgCl 2 1.2 μL, 1.5 μL of 2.5 mM dNTP Mixture, 1 μL of the bacterial solution as a template, 0.6 μL of primer F (10 μM), 0.6 μL of primer R (...
Embodiment 3
[0046] Example 3 Screening and Validation of Dominant Molecular Markers Specific to Bacterial Spot of Rice
[0047] According to the ePCR screening results of the specific primers specialized for rice bacterial leaf spot bacterium, adopt the genomic DNA of the test bacterial strain that has obtained, utilize the optimized PCR reaction system identical with embodiment 2 and 2% TBE agarose For the detection method of gel electrophoresis, 40 pairs of primers were selected to verify the tested strains.
[0048] After testing, we obtained 23 pairs of specific primers that can accurately amplify 10 strains of Bacteria sativa in all tested strains to a DNA fragment that matches the target size (see Table 6 for the fragment size). None of these specific primers could amplify bands in Xanthomonas oryzae and other reference strains (such as image 3 shown).
[0049] Table 6 The sequence numbers of such molecular markers in the sequence listing and the names of the corresponding molecu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com