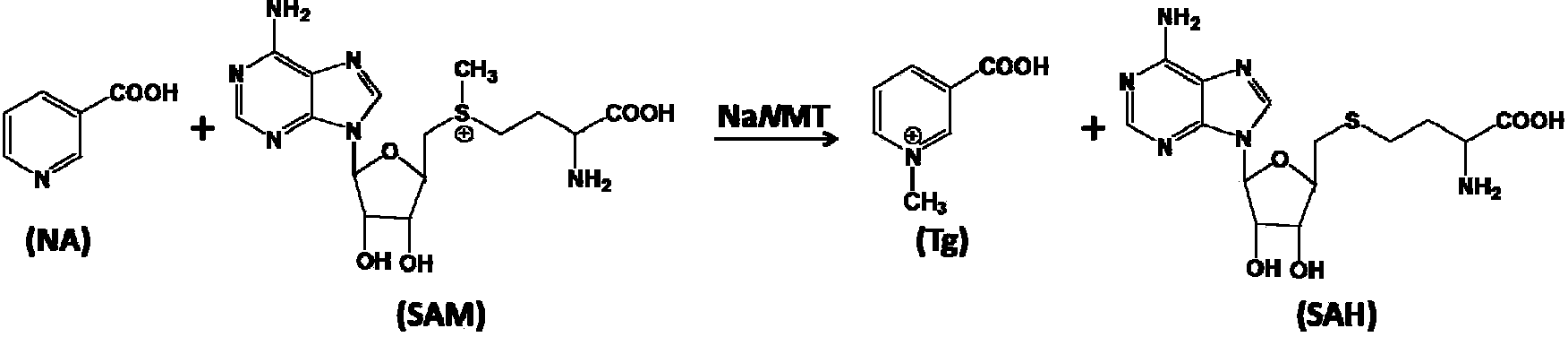
Soybean trigonelline synthase as well as coding gene and application thereof
A technology that encodes genes and trigonelline, which is applied in the fields of application, genetic engineering, plant gene improvement, etc., can solve the problems of slow research progress and achieve the effect of wide application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Embodiment 1, the acquisition of GmTS protein and its coding gene
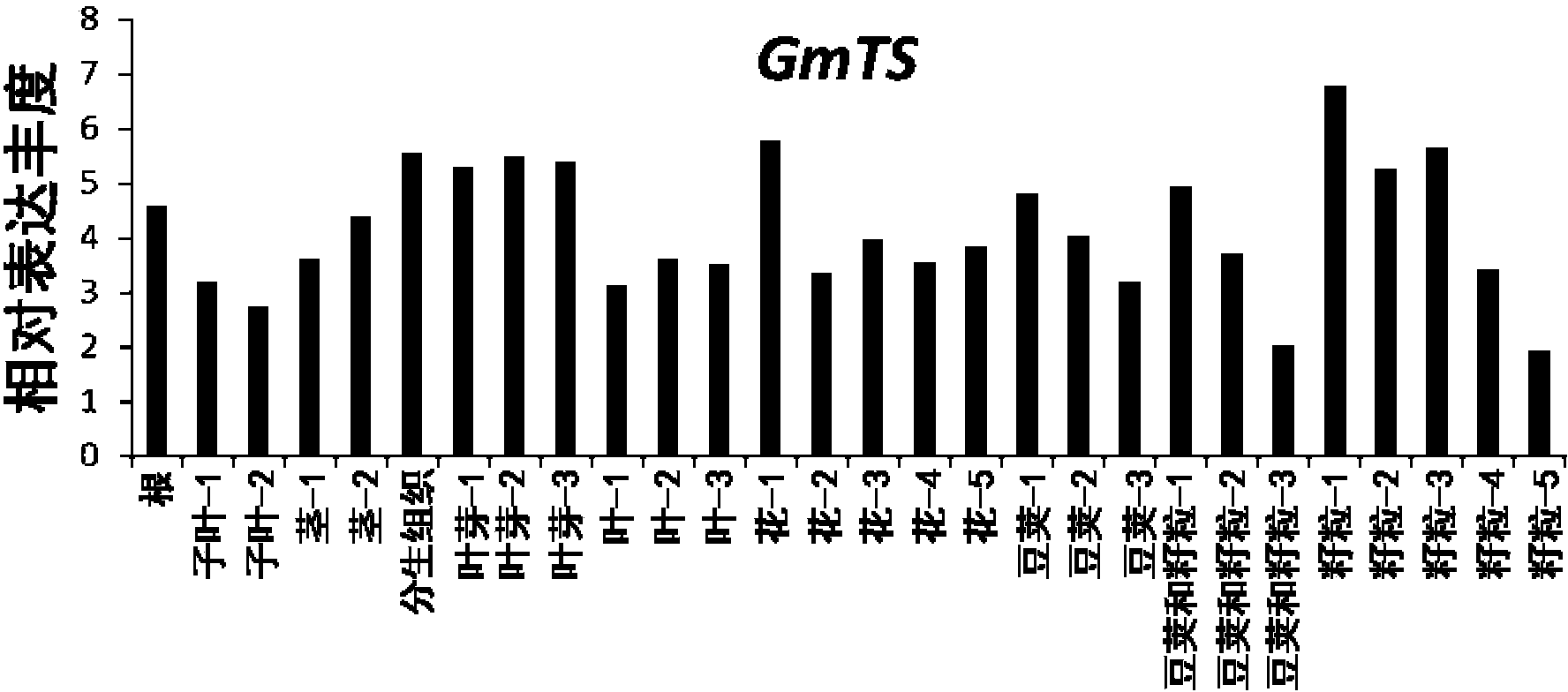
[0052] 1. The Trizol method was used to extract soybean (Glycine max cv Williams82) roots, cotyledons, stems, meristems, leaf buds, leaves, flowers, pods, pods and grains, and the total RNA of grains was extracted with GoScript TM ReverseTranscription System Reverse Transcription Kit were reverse transcribed into cDNA.
[0053] 2. Design and synthesize the following sequence:
[0054] GmTS F:5'-CCG GAATTC ATGGAGAAAGAGGAGAGCACGG-3' (SEQ ID No.1)
[0055] (The underlined sequence is the recognition site for EcoR I digestion)
[0056] GmTS R:5'-GC TCTAGA TCATTTCTGAAACTCAAGGACAGTGT-3' (SEQ ID No. 2)
[0057] (The underlined sequence is the Xba I restriction recognition site)
[0058] Actin F: 5'-ATCTTGACTGAGCGTGGTTATTCC-3' (SEQ ID No.3)
[0059] Actin R: 5'-GCTGGTCCTGGCTGTCTCC-3' (SEQ ID No.4)
[0060] 3. Use the cDNA of each soybean tissue in step 1 as a template, use Acitin F and Actin R as pri...
Embodiment 2
[0064] Embodiment 2, construction of recombinant plasmid pMal-GmTS
[0065] EcoR I and Xba I double-digest the DNA molecule shown in SEQ ID No.5 to obtain a gene fragment; EcoR I and Xba I double-enzyme digest the pMal-c2x vector to obtain a large vector fragment; connect the gene fragment to the large vector fragment, The recombinant plasmid was obtained, named pMal-GmTS, and pMal-GmTS was sent for sequencing, and the result was correct.
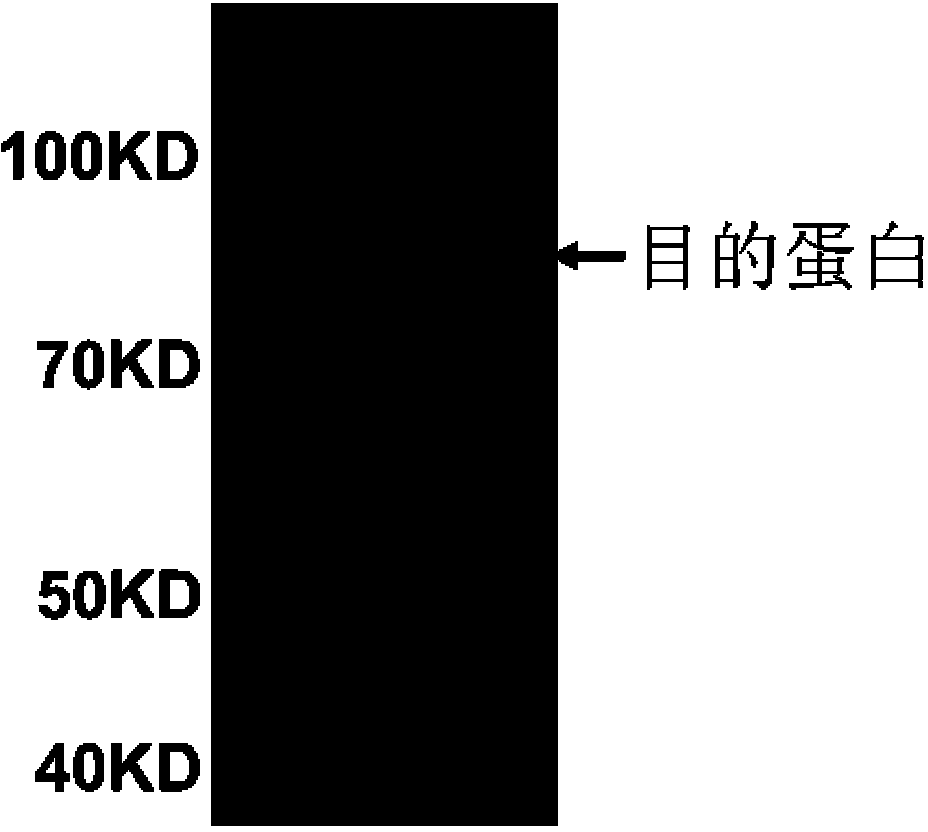
[0066] The structure of the recombinant plasmid pMal-GmTS is described as follows: between the EcoR I and Xba I restriction sites of the pMal-c2x vector, the 10th to 1077th nucleoside from the 5' end of SEQ ID No.5 is inserted The DNA molecule indicated by the acid, the plasmid expresses the GmTS-MBP fusion protein, and the expected molecular weight is about 80KD.
Embodiment 3
[0067] Embodiment 3, preparation of GmTS protein and enzyme activity analysis
[0068] 1. Construction of recombinant bacteria and control bacteria
[0069] The recombinant plasmid pMal-GmTS was introduced into Escherichia coli BL21(DE3)pLysS to obtain recombinant bacteria, and the pMal-c2x empty vector was introduced into Escherichia coli BL21(DE3)pLysS to obtain control bacteria.
[0070] 2. Preparation of GmTS protein
[0071] (1) Inoculate pMal-GmTS recombinant bacteria into LB Rich liquid medium (containing 100 μg / mL ampicillin), and culture at 37°C and 200 rpm until OD 600 =0.8, then add IPTG (to make its final concentration in the culture system 0.2g / L) and change the shaking culture condition to 16°C, and shake at 200rpm for 16 hours to obtain a culture solution.
[0072] (2) Centrifuge the culture solution to collect the bacterial cells.
[0073] (3) The cells were disrupted by ultrasonication, and the protein was purified by affinity chromatography using Amylose r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com