Method for biosynthesis of tyrosol in Escherichia coli and application of tyrosol
A technology of Escherichia coli and biosynthesis, applied in the field of bioengineering, can solve the problems of difficult industrialization, cumbersome process, low concentration, etc., and achieve the effect of important economic value and social benefit
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
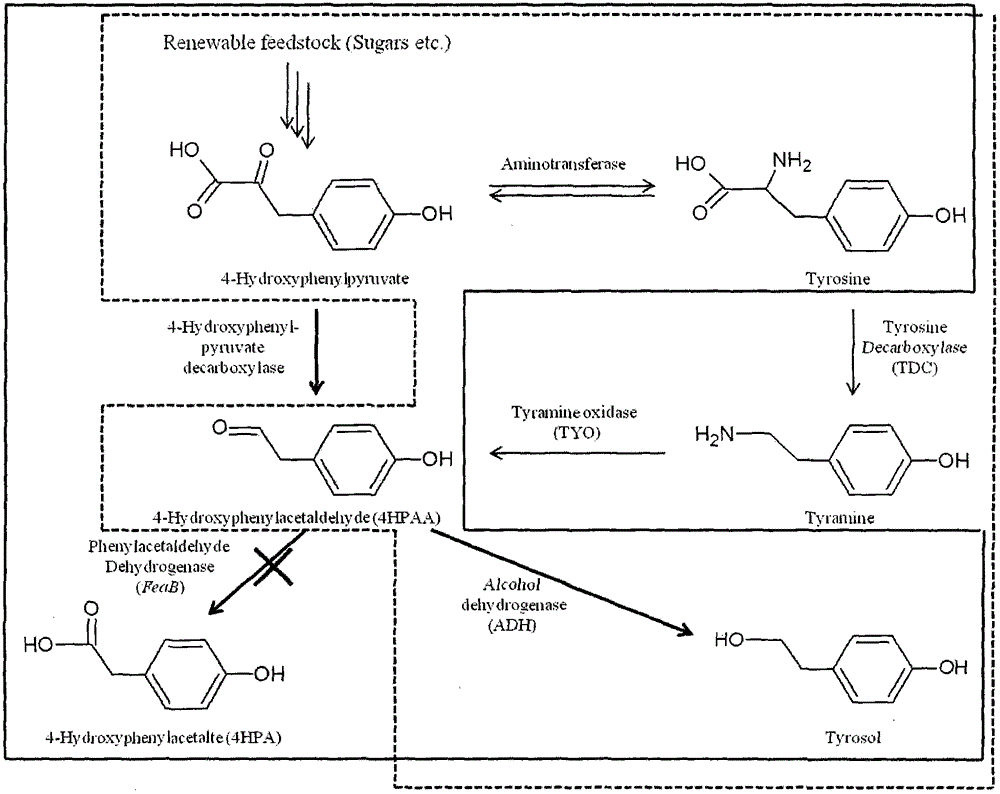
[0027] Example 1 Knockout of Escherichia coli phenylacetaldehyde dehydrogenase encoding gene FeaB
[0028] (1) Escherichia coli MG1655, plasmid pKD46 (temperature-sensitive type, containing exo, bet and gam genes regulated by the arabinose promoter, Amp r ), pKD4 (contains Kanna resistance gene with FRT sites at both ends, Kan r ), pCP20 (temperature-sensitive type, encoding FLP recombinase capable of recognizing FRT sites, Amp r ) are kept by the laboratory.
[0029] (2) Primer design and synthesis: According to the sequence information of FeaB in GenBank and Kan gene in plasmid pKD4, primers were designed and synthesized by Shenzhen Huada Gene Technology Co., Ltd. Primers FeaB-5FPL / FeaB-3RPL are used to amplify the Kan gene to replace the coding region of the FeaB gene; primers MG1655-5FP / MG1655-3RP are primers for identification of recombinant bacteria after FeaB gene knockout, and the two primers are located on the left and right of the target sequence region sides.
...
Embodiment 2
[0037] Example 2 Amplification of yeast 4-hydroxyphenylpyruvate decarboxylase encoding gene ARO10 and expression vector construction and transformation
[0038](1) According to the gene sequence of yeast 4-hydroxyphenylpyruvate decarboxylase coding gene ARO10 provided in NCBI, primers ARO10-5FP and ARO10-3RP were designed, and the genomic DNA of Saccharomyces cerevisiae S288C was used as a template for PCR, amplified to obtain the ORF of ARO10, such as image 3 shown. The primer sequences are as follows (the restriction site is indicated by the underline):
[0039]
[0040] reaction system:
[0041]
[0042]
[0043] Reaction program: initial denaturation at 98°C for 30 sec; 30 cycles of 98°C for 8 sec, 55°C for 30 sec, and 72°C for 1 min; final extension at 72°C for 10 min.
[0044] (2) The ARO10 and plasmid pTrcHisB amplified in step (1) were digested by SacI and NcoI respectively, recovered, and the digested ARO10 was connected into pTrcHisB to obtain the ARO10 ...
Embodiment 3
[0046] The fermentation culture of embodiment 3 recombinant strains
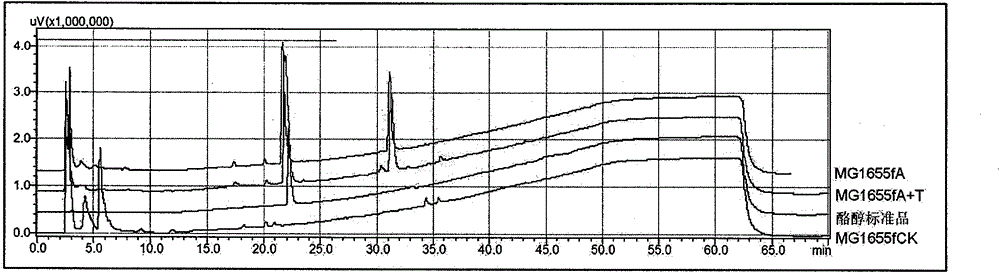
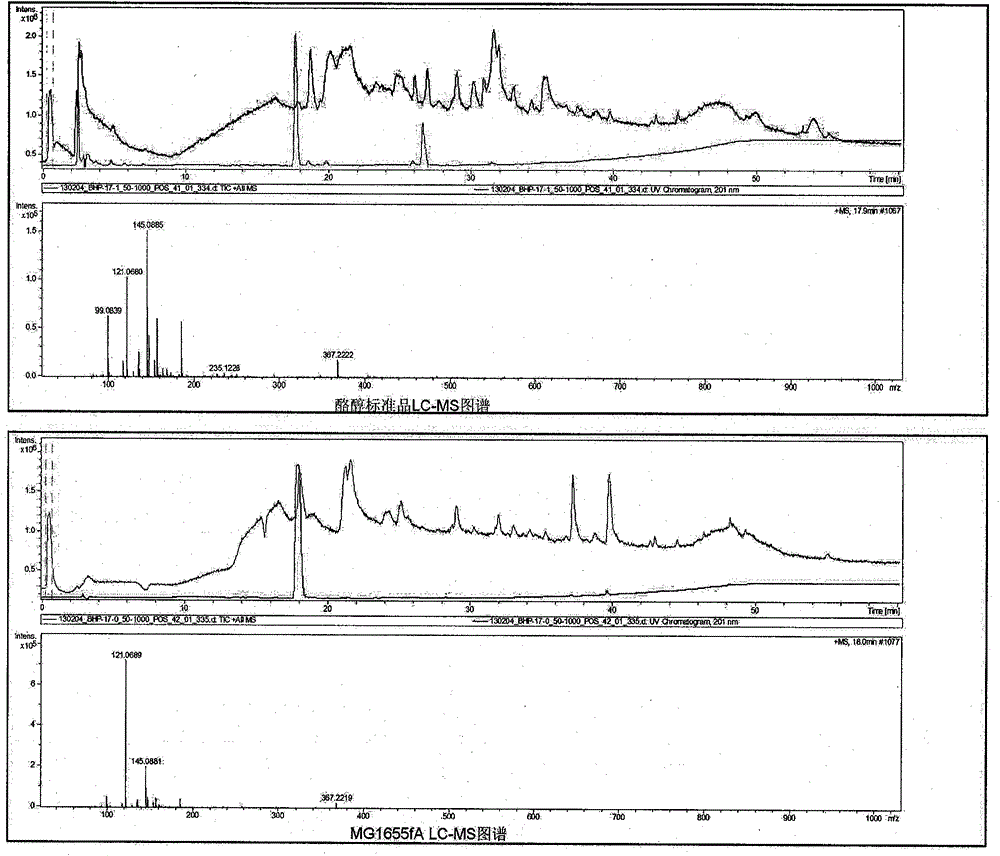
[0047] Shake flask fermentation: the recombinant strain was cultured overnight at 37°C in 2mL LB medium, then transferred to 50mL fresh LB medium at 1% inoculum size and cultured at 37°C until the OD600 was about 0.6, adding IPTG with a final concentration of 0.5mM to induce After culturing at 30°C for 5 hours, the cells were collected, washed once with M9Y (M9 medium added with 0.125% Yeast extract), suspended in 50 mL of M9Y medium containing 1% glucose and cultured at 30°C for 20 hours. There were three groups of samples, MG1655fCK (control group), MG1655fA+T (experimental group, fed with 1 mM tyrosine), and MG1655fA (experimental group, not fed with tyrosine). The specific formula of M9Y medium is as follows:
[0048]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com