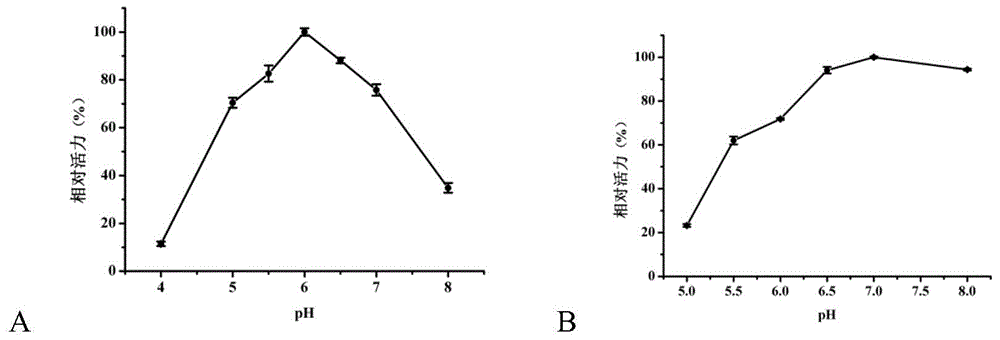
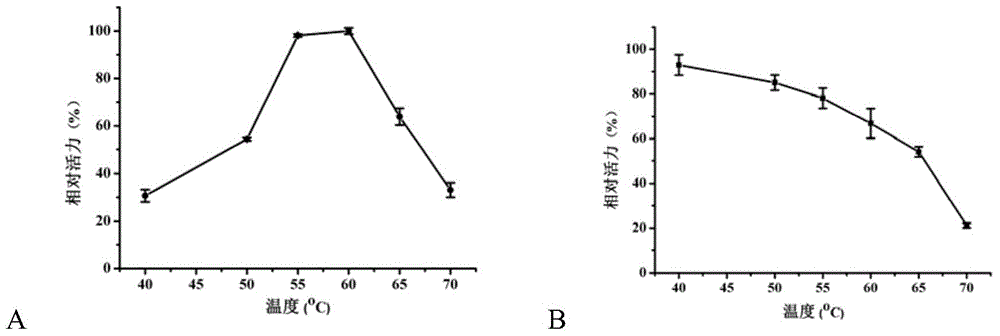
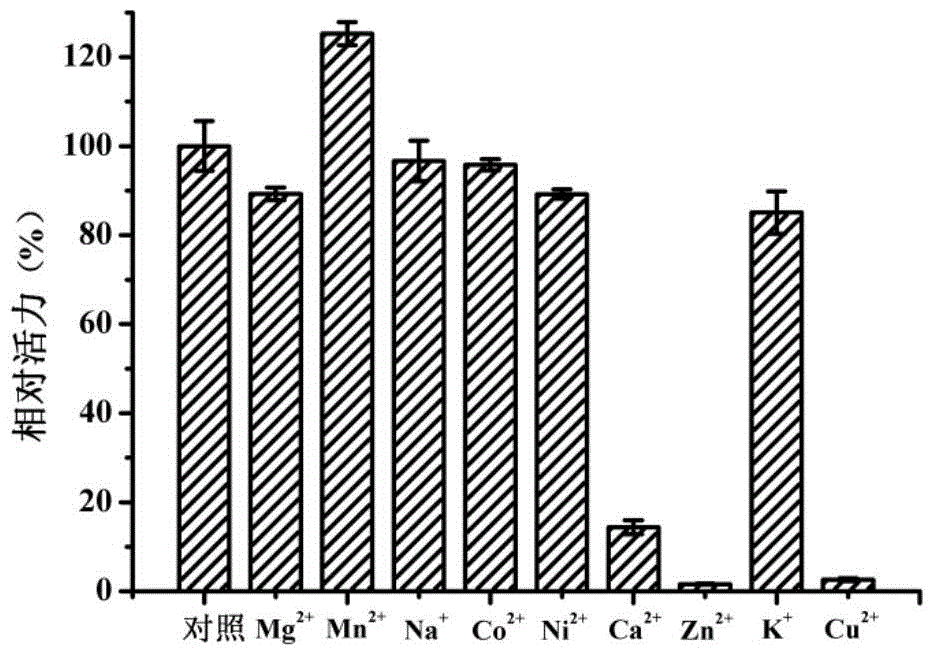
Beta-galactosidase and application thereof
A technology of galactosidase and nucleotide sequence, applied in the fields of application, glycosylase, enzyme, etc., can solve problems such as protein sequence differences, and achieve the effect of high pH stability and thermal stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: PCR amplification of Bacillus coagulans NL01β-galactosidase gene.
[0027] Genomic DNA of Bacillus coagulans NL01 was extracted using a genome extraction kit from Promega.
[0028] Design primers to introduce the EcoR I and Sal I restriction sites that can be inserted into the pETDuet-1 plasmid (Novagen), the sequences are as follows:
[0029] gal.f: 5'-GAATTCGATGTTAAAAAAACAAGAAAAATTTTATTATG-3'
[0030] gal.r: 5'-GTCGACCTATTTTTCAATTACCTGCAAAATTTTCA-3'
[0031] Using the extracted Bacillus coagulans NL01 genomic DNA as a template, the above primers were used for PCR amplification. The total PCR amplification system was 100 μL, containing 64 μL of ultrapure water, 20 μL of 5×FastPfu PCR Buffer, 0.25 mmol / L of dNTPs, 0.2-0.4 μmol / L of each primer, 50-200 ng of template, and FastPfu DNA Polymerase 5U from TransGen Company. PCR amplification conditions were: pre-denaturation at 95°C for 2 minutes, 30 cycles according to the following parameters (denaturation at...
Embodiment 2
[0032] Example 2: Cloning and sequence determination of PCR products.
[0033] The PCR product of Example 1 was recovered and connected to the pEasy-Blunt cloning vector of TransGen Company, transformed into Escherichia coli Mach1-T1 competent cells, coated with ampicillin plate, screened for positive recombinant strains and sequenced. The sequencing was completed by Beijing Liuhe Huada Gene Company, and the sequence analysis of the sequencing results showed that the PCR product was 1998bp, encoding 665 amino acids, as shown in SEQ ID No:1.
Embodiment 3
[0034] Example 3: Expression and purification of Bacillus coagulans NL01β-galactosidase gene in Escherichia coli
[0035] The correct recombinant strain screened in Example 2 was inoculated into 5 mL of LB liquid medium containing 100 μg / mL ampicillin, cultured on a shaker at 37° C. for 8 to 12 hours, and the recombinant plasmid was extracted, digested with EcoR I and Sal I for 1 to 3 Hour. The pETDuet-1 plasmid expression vector was treated with the same digestion method, and the digested product was separated and purified by agarose gel electrophoresis. Use T4 DNA ligase from NEB Company for 1 hour at 22°C and then use CaCl 2 coli BL21 (DE3) competent cells were transformed by the method, ampicillin plates were spread, positive recombinant strains were screened and plasmids were extracted, EcoR I and Sal I double enzyme digestion identified the screened recombinant plasmids. The strain containing the correct recombinant plasmid was named E.coli BL21(pETDuet-gal). At the s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com