A screening method for nucleic acid aptamers specifically binding to alpha-fetoprotein
A nucleic acid aptamer and alpha-fetoprotein technology, applied in the field of analytical chemistry, can solve the problems of immunogenicity and high cost of antibodies
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1 Screening of nucleic acid aptamers
[0026] 1. Instruments and materials: P / ACEMDQ capillary electrophoresis instrument (Beckman-Coulter, USA), equipped with diode array (PDA) and laser-induced fluorescence (LIF) detectors; PVA-coated elastic quartz capillary column with a total length of 40cm and an effective length of 30cm , with an inner diameter of 50 μm and an outer diameter of 360 μm.
[0027] 2. Electrophoresis buffer preparation: use ultrapure water to prepare 30mM NaH 2 PO 3 , adjusted the pH value to 7.5 with 1M NaOH solution, and filtered through a 0.45 μm membrane filter.
[0028] 3. Design and synthesize a random oligonucleotide library with a full length of 75 bases, the sequence is shown in SEQ ID NO.1, 5'-GTGACGCTCCTAACGCTGAC-35N-CCTGTCCGTCCGAACCAATC-3', and the two ends are fixed with 20 bases respectively Sequence, the middle 35 bases are random sequences.
[0029] 4. Synthesize upstream primers and downstream primers for PCR amplificat...
Embodiment 2
[0038] Cloning and sequencing of embodiment 2 nucleic acid aptamers
[0039] Prepare the following solutions in a microcentrifuge tube, the total volume is 5 μl: pMD19-TVector 1 μl, oligonucleotide PCR product 0.2 pmol, ddH 2O supplemented to 5 μl. Add 5 μl of SolutionI. React at 16°C for 30 minutes. The whole amount (10 μl) was added to 100 μl Trans5α competent cells, and placed in ice for 30 minutes. After heating at 42°C for 45s, place in ice for 1min. Add 890 μl LB medium, shake and culture at 37°C for 60min. Culture on LB agar plate medium containing X-Gal, IPTG and Amp at 37°C to form a single colony. Select white monoclonal colonies and put them into 2ml LB (Amp80μg / ml) liquid medium, shake and culture at 200rpm, 37°C for 16h, and submit the bacterial liquid to Shanghai Sangon for sequencing.
Embodiment 3
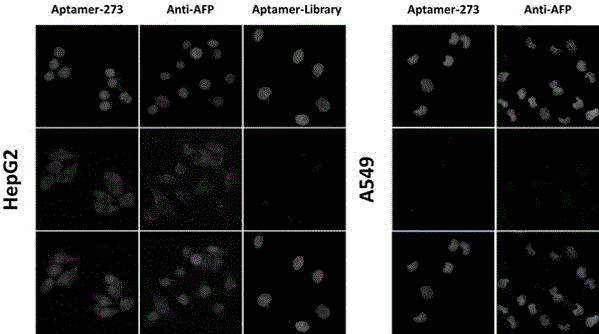
[0040] Example 3 Verification of nucleic acid aptamers
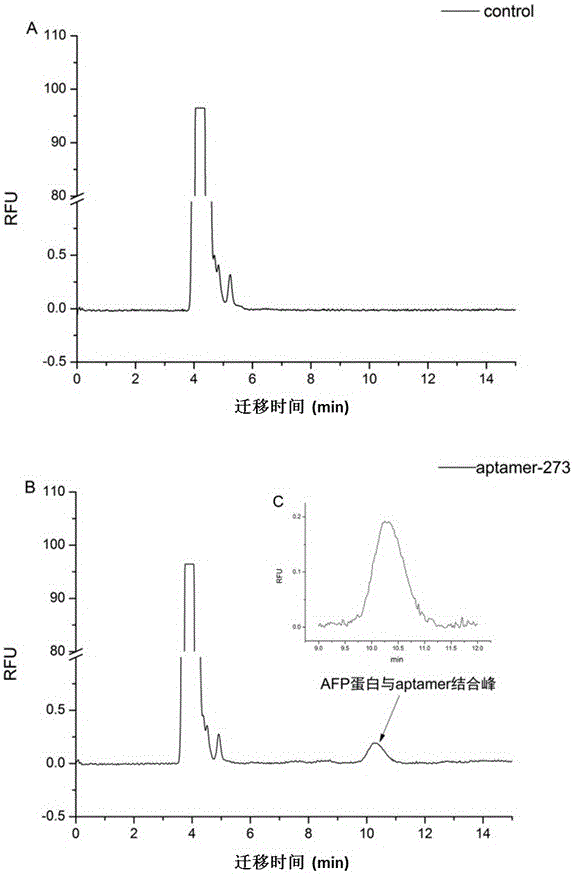
[0041] 83 oligonucleotide sequences were obtained by sequencing. After sorting out the sequencing results and predicting the structure, 14 sequences with representative structures were selected for capillary electrophoresis analysis, and one sequence with high specificity, aptamer-273, was selected.
[0042] The sequence of Aptamer-273 is shown in SEQ ID NO.4.
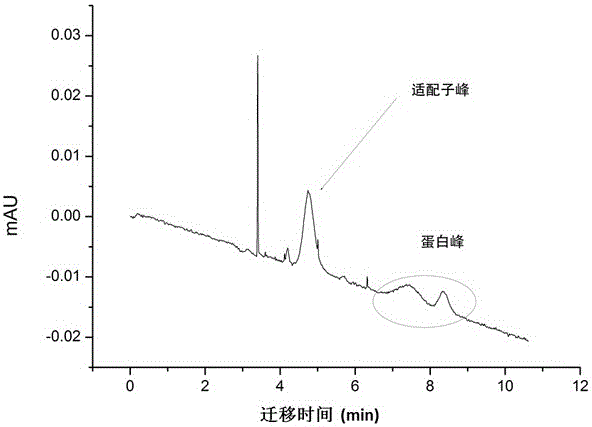
[0043] Investigation 1: Capillary electrophoresis detection of binding specificity between fluorescently labeled aptamer-273 and AFP protein in this example.
[0044] Electrophoresis buffer is 30mMNaH 2 PO 3 , pH7.5, AFP concentration is 0.1mg / ml, aptamer concentration is 10μM. The inner diameter of PVA-coated capillary column is 50 μm, the total length is 40 cm, and the effective length is 30 cm; the separation voltage is -8 kV, the pressure is 34 mbar; the temperature is 25 ° C; the pressure is 34 mbar, and the mixture of fluorescent (FAM)-labeled aptamer...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com