A method of production of 2,4-dihydroxybutyric acid
A technology of dihydroxybutyric acid and malic acid, applied in biochemical equipment and methods, carbon-carbon lyase, enzymes, etc., can solve problems such as uncertainty
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
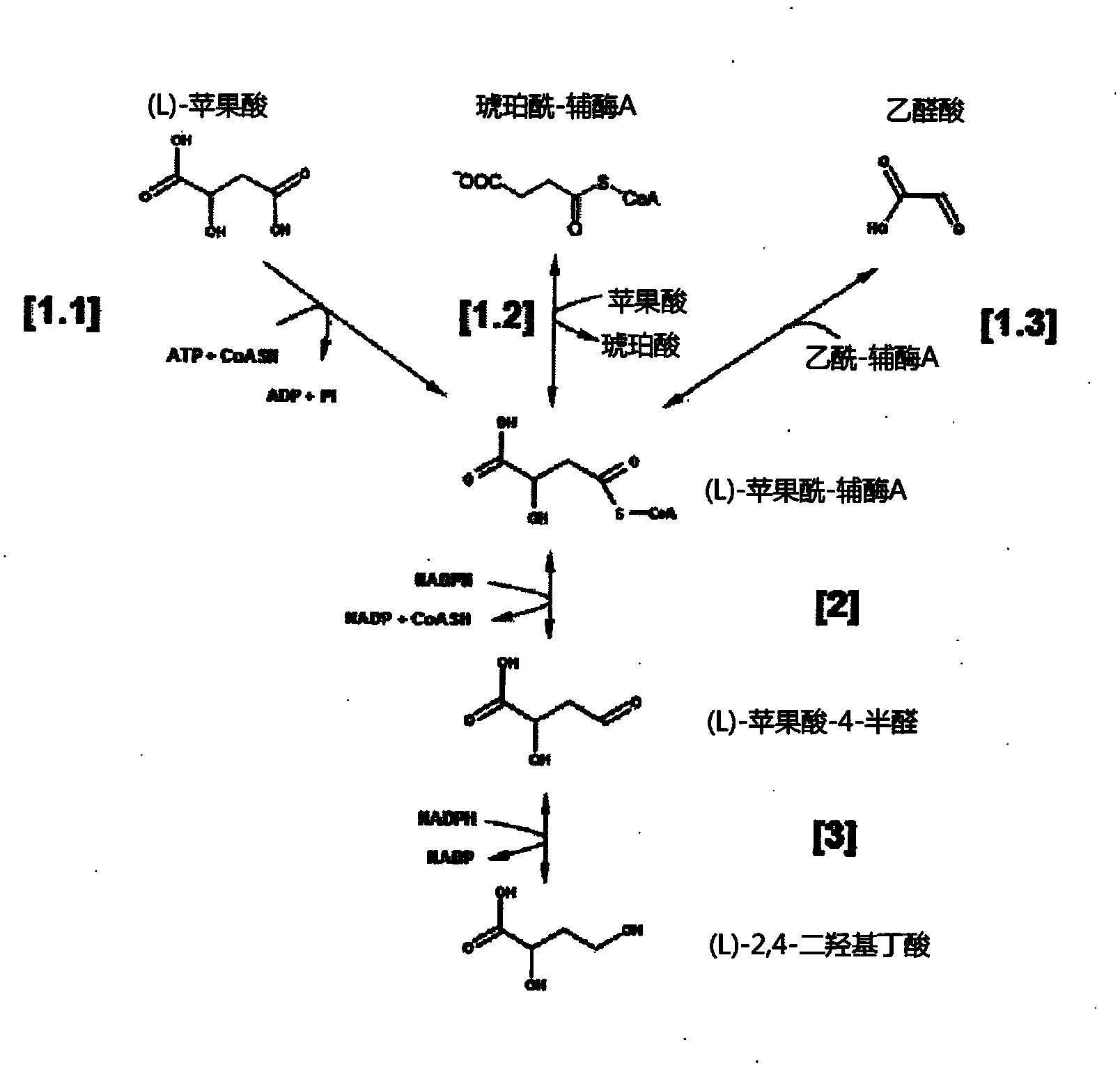
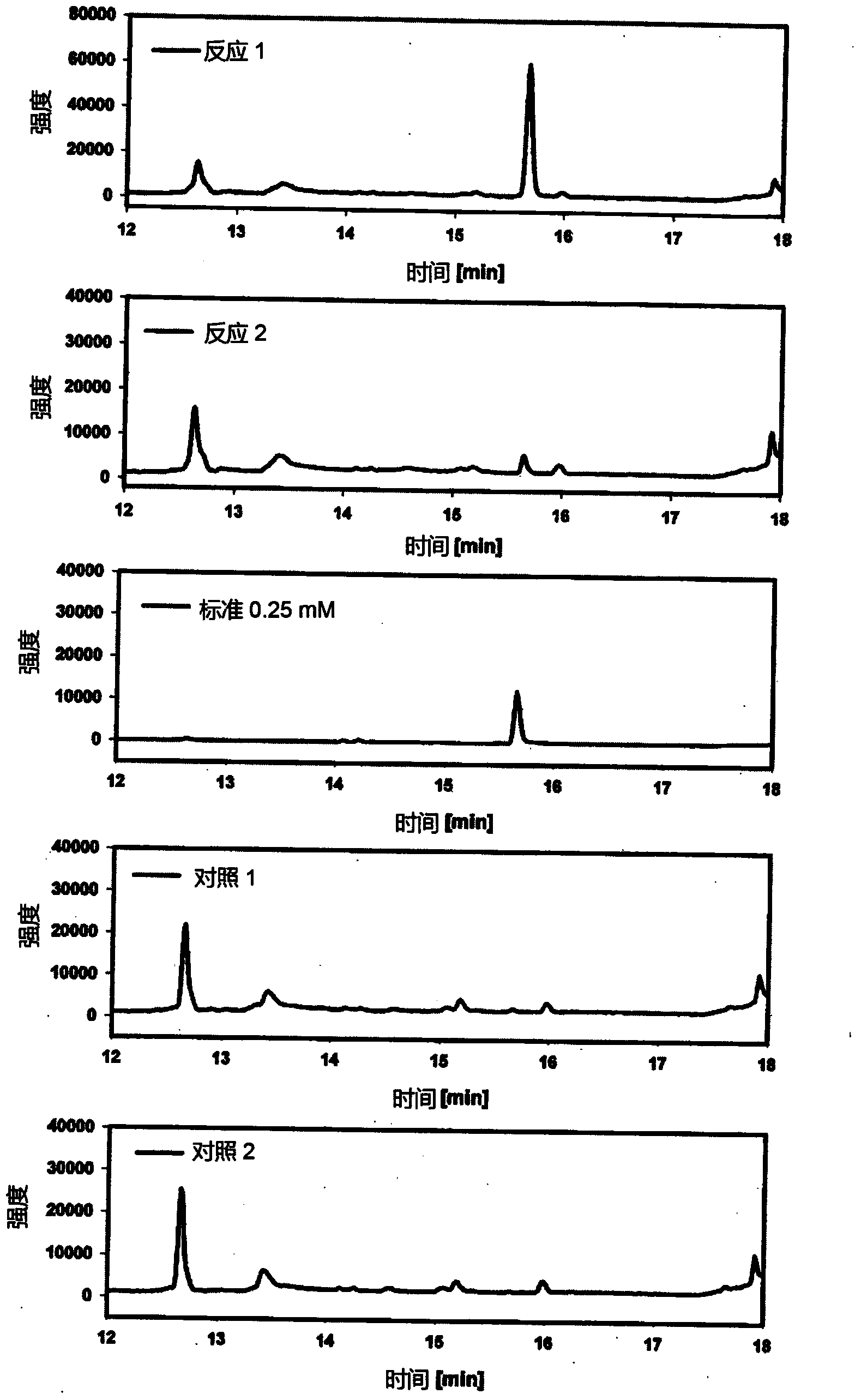
[0102] Example 1: Demonstration of malyl-CoA lyase activity
[0103] Construction of plasmids containing wild-type genes encoding malyl-CoA lyase: Encoding of malyl-CoA lyase in outer chain Methylobacterium (Arps et al., 1993) and Rhodobacter capsulata (Meister et al., 2005) The DNA sequence of the mcl gene was optimized for expression in E. coli using GENEius software (Eurofins). Optimized sequence by Eurofins MWG For synthesis, NheI and EcoRI were added at restriction sites upstream of the start codon and mcl was added downstream of the stop codon, which enabled clones of synthetic DNA fragments to directly enter pET28a+ vector (Novagen) using T4 DNA ligase (Biolabs). Bound products were expanded by transfection into E. coli DH5α cells, and the plasmids pET28-Mex-mcl (expressing malyl-CoA lyase from outer chain Methylobacterium) and pET28-Mex were isolated using standard genetic protocols (Sambrook et al., 1989). Rca-mcl (from M. capsulatus expressing malyl-CoA lyase). N...
Embodiment 2
[0118] Example 2: Demonstration of Malyl-CoA Reductase Activity
[0119] Construction of plasmids containing wild-type genes encoding malyl-CoA reductase and succinyl-CoA reductase: encoding of malyl-CoA reductase mcr in the hyperthermophilic archaea str 7 (Alber et al., 2006) The DNA sequence of the gene was optimized for expression in E. coli using the GENEius software (Eurofins). Optimized mcr sequence and native DNA sequence of the sucD gene encoding succinyl-CoA reductase in Porphyrophyllum gingivalis W83 via Eurofins MWG For synthesis, NheI and EcoRI were added upstream of the start codon and mcl downstream of the stop codon, respectively, which enabled direct clones of synthetic DNA fragments into pET28a+ vector (Novagen) using T4 DNA ligase (Biolabs) . Bound products were expanded by transshipment into E. coli DH5α cells, and the plasmids pET28-St-mcr (expressing malyl-CoA reductase from hyperthermophilic archaea) and pET28 were isolated using standard genetic proto...
Embodiment 3
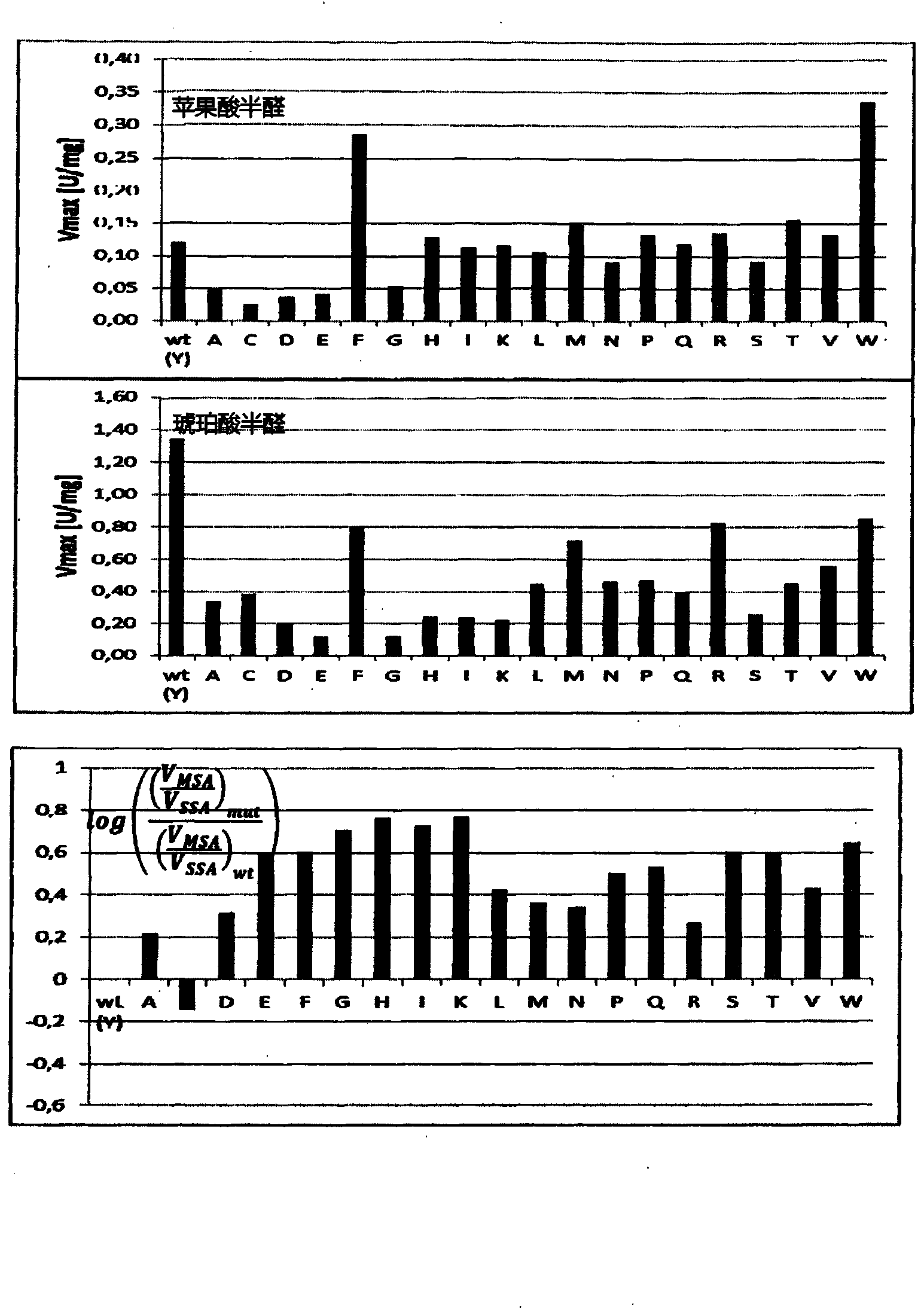
[0141] Example 3: Demonstration of DHB reductase activity
[0142] To identify suitable 2,4DHB reductases, β-hydroxycarboxylic acid dehydrogenases of different biological origins were tested for their ability to reduce malate semialdehyde. The enzymes tested were: methylbutyraldehyde reductase Ypr1 from Saccharomyces cerevisiae (Ford & Ellis, 2002) (SEQ ID No.14), 4-hydroxybutyrate dehydrogenase from Porphyrophyllum gingivalis, 4hbdh (SEQ ID No.187 ), alcohol dehydrogenase YqhD from Escherichia coli (SEQ ID no.185), and succinate semialdehyde reductase Ms-Ssr from Metallococcus (Kockelkorn & Fuchs, 2009) (SEQ ID No.16). Use the primers listed in Table 5 to amplify the genes YPR1, 4hbdh, yqhD, and Ms-SSR and clone them into the vector pET28 (see Table 5 for restriction enzymes) to obtain plasmids pET28-Sce-YPR1 and pET28-Pgi-4hbdh respectively , pET28-Eco-yqhd and pET28-Mse-SSR. The protein was expressed and purified as described in Example 1.
[0143]
[0144] table 5:...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com