Method for optimizing signal peptide and improving exocytosis expression of trypsin
A technology of trypsin and trypsin gene, applied in the direction of microorganism-based methods, biochemical equipment and methods, peptidase, etc., can solve the problems of low trypsin activity and low protein expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
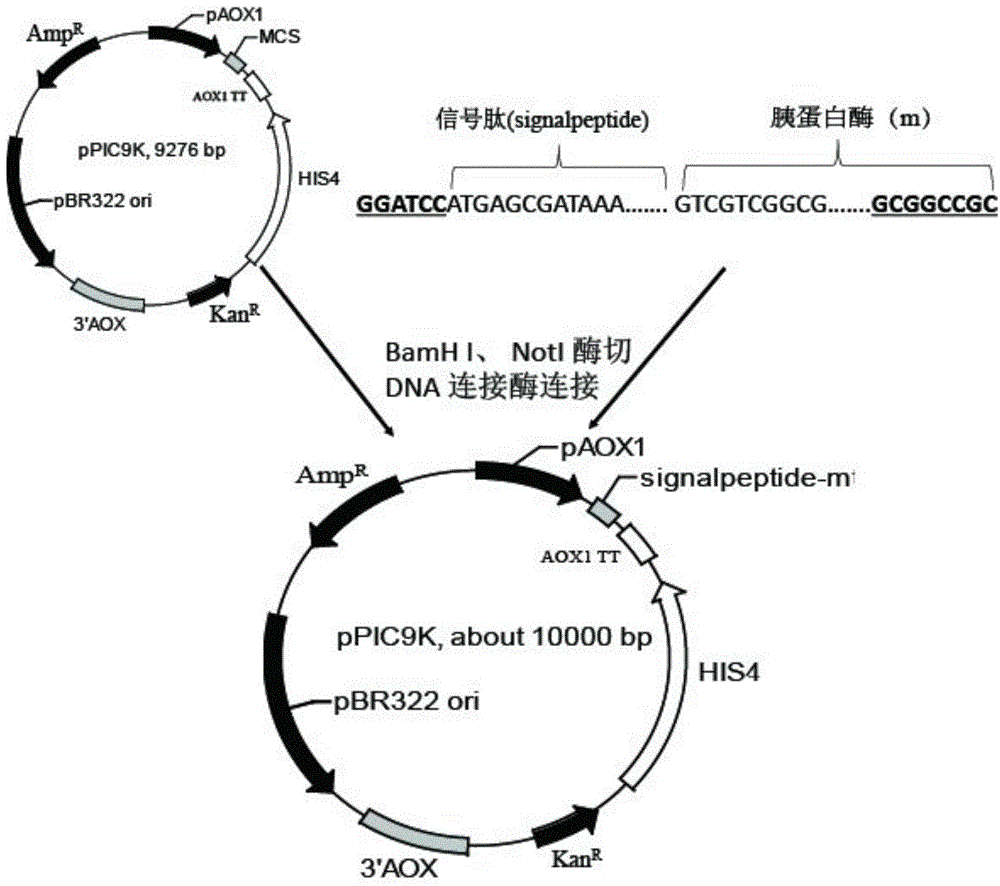
[0025] Example 1 Construction of recombinant plasmid
[0026] The trypsin gene of Streptomyces griseus (Gen Bank Accession No. M64471) is 672 bp in length. Design primers based on the nucleic acid sequences of the eight signal peptides. After two-step PCR, the signal peptide sequence and the gene sequence shown in SEQ ID NO.9 are homogenized to obtain the fusion gene fragment (using the trypsin gene shown in SEQ ID NO.9 as the template, the F2 and R primers are used for PCR and then the column is recovered. Then use the F1, R primer PCR column to recover the product to obtain the fusion fragment. The F1, F2, and R primer sequences are shown in SEQ ID NO. 10, SEQ ID NO. 11, and SEQ ID NO. 12 respectively), according to figure 1 In the construction method shown, the fusion fragment and pPIC9K plasmid were digested with NotI and BamH I, and then ligated with T4 ligase overnight at 16°C after purification. The ligation product chemically transforms JM109 competent cells. The transf...
Embodiment 2
[0027] Example 2 Construction of trypsin-producing yeast engineering bacteria
[0028] The recombinant plasmid containing the recombinant gene with signal peptide added before the trypsin gene was linearized with Sal I, and electroporation was used to transform pichia pastoris GS115 competent cells. The specific method is as follows:
[0029] 1) Inoculate pichia pastoris GS115 activated on YPD plates in 25mL / 250mL Erlenmeyer flasks and culture overnight at 30℃; 1% inoculate the above-mentioned culture solution in 50mL / 500mL Erlenmeyer flasks, culture cell concentration OD600 is 1.3~1.5;
[0030] 2) Centrifuge at 5000r / min for 10min at 4℃ to collect the bacteria, and suspend the cells in 50mL and 25mL sterile water respectively;
[0031] 3) Resuspend the above cells in 5mL 1M sorbitol, centrifuge at 5000r / min for 10min at 4℃ to collect the bacteria;
[0032] 4) Resuspend the above cells in 500μL of 1M sorbitol, and aliquot 80μL / 1.5mL EP tubes for electrotransformation of competent cells;...
Embodiment 3
[0037] Example 3 Construction of recombinant bacteria
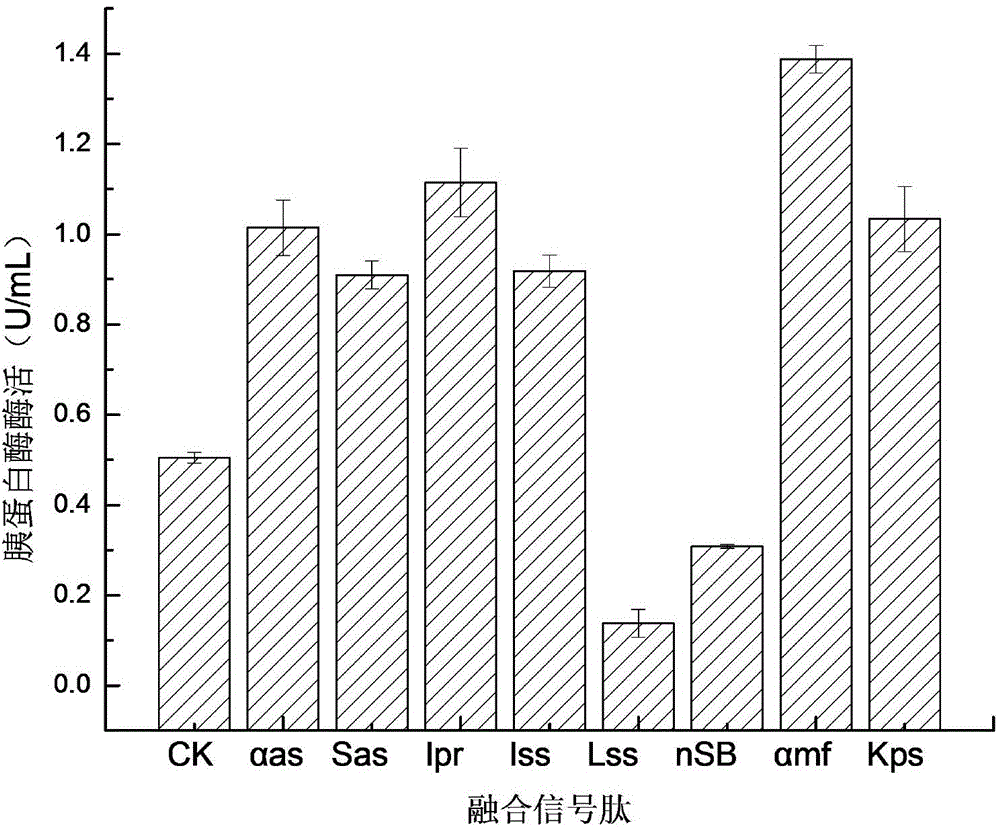
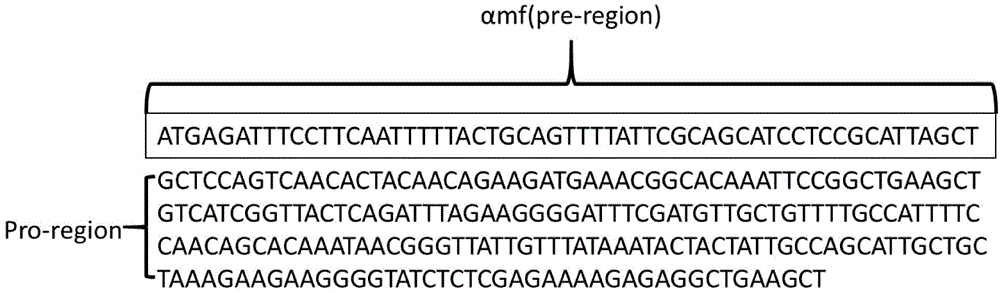
[0038] Using a two-step PCR method similar to Example 1, the nucleotide sequences such as the signal peptides of SEQ ID NO.1 to SEQ ID NO.8 (named as αas, Sas, Ipr, Iss, Lss, nsB, αmf, Kps) before fusion to the trypsin gene shown in SEQ ID NO.9, 8 recombinant genes αas-mt, Sas-mt, Ipr-mt, Iss-mt, Lss-mt, nsB-mt, αmf-mt, Kps-mt; respectively ligate the recombinant genes to the Pichia expression vector to obtain a recombinant plasmid; respectively electrotransform the recombinant plasmids into Pichia pastoris GS115 to obtain a recombinant yeast engineering strain.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com