Fusion protein capable of improving dammarendiol conversion efficiency and construction method
A technology for fusion protein and dammarenediol, which is applied in the field of fusion protein and construction, and can solve the problem of low conversion efficiency of dammarenediol
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Construction of Saccharomyces cerevisiae cells for the synthesis of dammarenediol
[0035] 1. Module construction
[0036] According to the amino acid sequence of dammarenediol synthase, codon optimization for Saccharomyces cerevisiae was carried out, and then the gene DS encoding dammarenediol synthase was obtained by chemical synthesis method (synthesized by Jinweizhi Biotechnology Co., Ltd.) as SEQ ID NO.4; Saccharomyces cerevisiae endogenous tHMG1 (SEQ ID NO.5), erg1 (SEQ ID NO.6), and promoters PGK1p (SEQ ID NO.7), TEF1p (SEQ ID NO.8), TDH3p (SEQ ID NO.9) and the terminator CYC1t (SEQ ID NO.10), ADH1t (SEQ ID NO.11), ADH3t (SEQ ID NO.12) are all from Saccharomyces cerevisiae w303-1a genome; selection marker gene leu2 is from Plasmid prs405 (ATCC, USA), his3 was from plasmid pxp320 (purchased from Addgene, Inc.WWW.addgene.org).
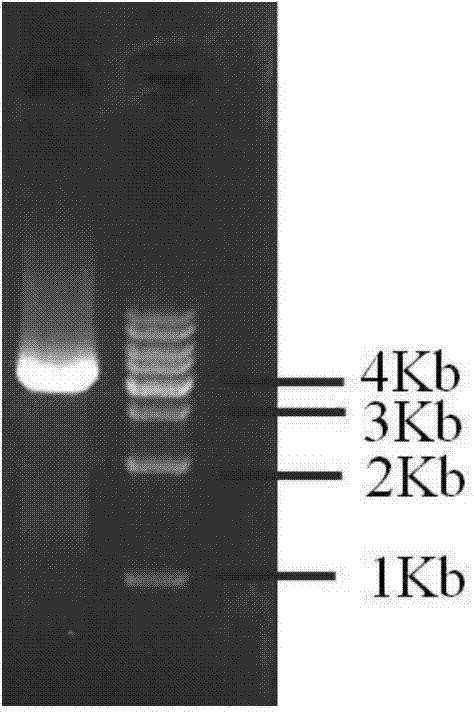
[0037] Using the Saccharomyces cerevisiae W303-1a (USA, ATCC) genome as a template, PGK1p-DS-F (SEQ ID NO.13) (and PGK1p-DS-R (SEQ ID NO...
Embodiment 2
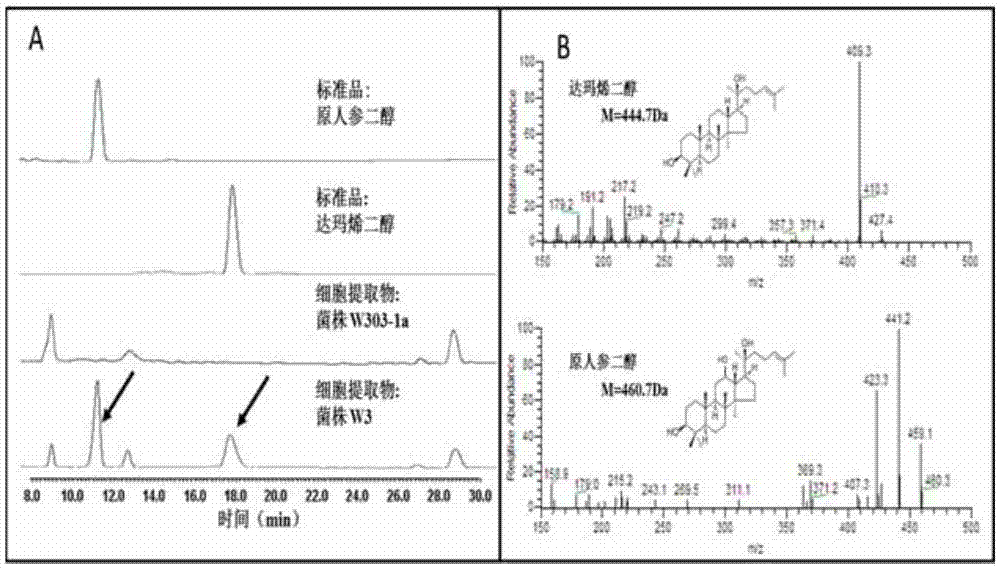
[0048] Example 2, the construction of synthetic protopanaxadiol Saccharomyces cerevisiae cells W3 and W3plus
[0049] According to the amino acid sequence of protopanaxadiol synthase and cytochrome-NADPH-reductase 1 in Arabidopsis, codon optimization for Saccharomyces cerevisiae was carried out, and then the code was obtained by chemical synthesis (synthesized by Jinweizhi Biotechnology Co., Ltd.) The gene PPDS gene of Panaxadiol synthase in Panax ginseng is SEQ ID NO.3 in the sequence listing, and the AtCPR1 gene in Arabidopsis thaliana is the gene sequence SEQ ID NO.1 in the sequence listing. TDH3p, PGK1p and terminator CYC1t, ADH3t are all from S. cerevisiae w303-1a genome; selection marker gene ura3 is from plasmid pxp218 (Addgene, Inc.WWW.addgene.org).
[0050] Using the Saccharomyces cerevisiae W303-1a genome as a template, TDH3p-AtCPR1-F (SEQ ID NO.43) and TDH3p-AtCPR1-R (SEQ ID NO.44) and AtCPR1-ADH3T-F (SEQ ID NO.47) and AtCPR1-ADH3T-R (SEQ ID NO.48) was used as a pr...
Embodiment 3
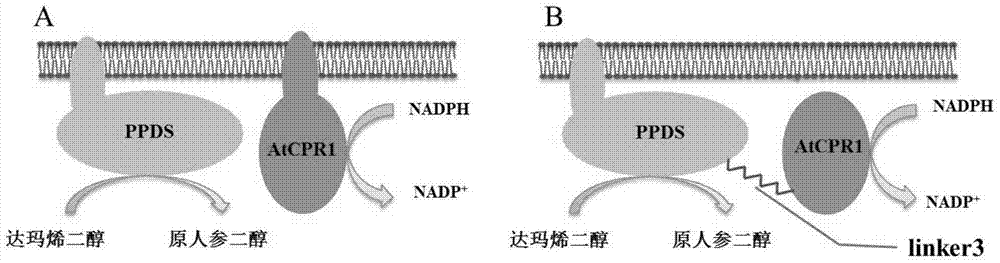
[0054] Example 3. Construction of a fusion protein that can improve the conversion efficiency of dammarenediol
[0055] According to the amino acid sequence of the protopanaxadiol synthase gene PPDS and the cytochrome-NADPH-reductase 1 gene AtCPR1 in Arabidopsis thaliana, codon optimization for Saccharomyces cerevisiae was carried out, and then chemically synthesized (Jinweizhi Biotechnology Co., Ltd. Synthesis) to obtain gene fragments. The PPDS gene in ginseng is shown in SEQ ID NO.3, and the AtCPR1 gene in Arabidopsis is shown in SEQ ID NO.1; the first 138 bases at the 5' end of the AtCPR1 gene in Arabidopsis are excised to obtain SEQ ID NO. 2 sequence shown. Both the endogenous promoter PGK1p (SEQ ID NO.7) and the endogenous terminator ADH3t (SEQ ID NO.12) are from the genome of Saccharomyces cerevisiae w303-1a; the selection marker gene ura3 is from the plasmid pxp218.
[0056]The two proteins are linked by a base sequence encoding the polypeptide GGG. In this example, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com