Wheat salt-tolerant gene TaBASS2 and application thereof
A wheat salt tolerance gene and gene technology, applied in application, genetic engineering, plant genetic improvement and other directions, to achieve the effect of improving salt tolerance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Example 1, Cloning of Shanrong No. 3 Wheat Salt Tolerance Gene Allene Oxide Reductase Gene TaBASS2
[0020] 1.1 Processing of plant material
[0021] 1) Vernalization of wheat seeds (Shanrong No. 3) at 4°C for 20 days
[0022] 2) Treat the seeds with 70% alcohol for 2-3 minutes.
[0023] 3) Discard the alcohol, wash with sterile water for 3-5 times, shake and mix well each time.
[0024] 4) Soak the seeds in sterile water, avoid light, incubate overnight at 25°C, 40-60rpm / min, on a shaking table.
[0025] 5) Place the seeds face up on filter paper fully moistened with sterile water, and germinate in the dark.
[0026] 6) After 3 days, the seeds were transferred to the culture basket, 1 / 2 Hoagland, hydroponics, placed at 23°C, and grown to the stage of two leaves and one heart in the long-day culture room.
[0027] 1.2 Wheat RNA extraction
[0028] 1) Weigh the cryogenically frozen 30-50 mg RNA extraction sample and quickly transfer it to a mortar pre-cooled with li...
Embodiment 2
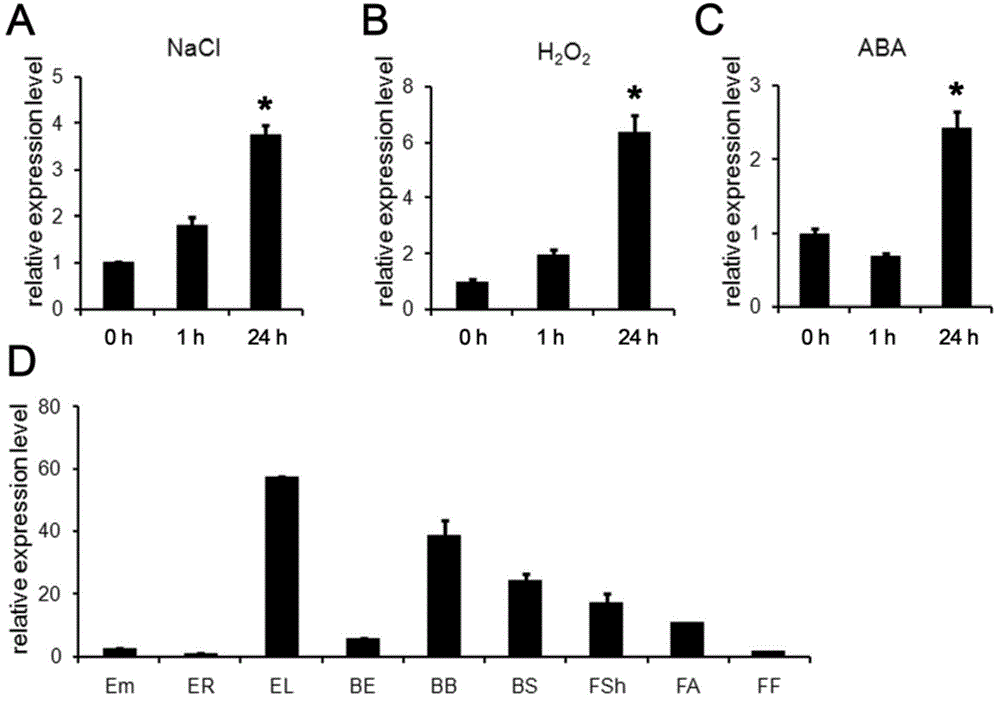
[0060] Embodiment 2, expression analysis of TaBASS2
[0061] 2.1 Extraction of RNA under stress
[0062] The seeds of Shanrong 3 and Jinan 177 germinated normally. When the Hoagland culture medium was cultured to the stage of two leaves and one heart (about 3 weeks), the treatments of drought (18% PEG), salt stress (200mM NaCl), cold and ABA began. After treatment for different time, the seedling root and leaf RNA was extracted by Trizol method as above.
[0063] 2.2 Reverse transcription to obtain cDNA
[0064] Reverse transcription generated cDNA as above.
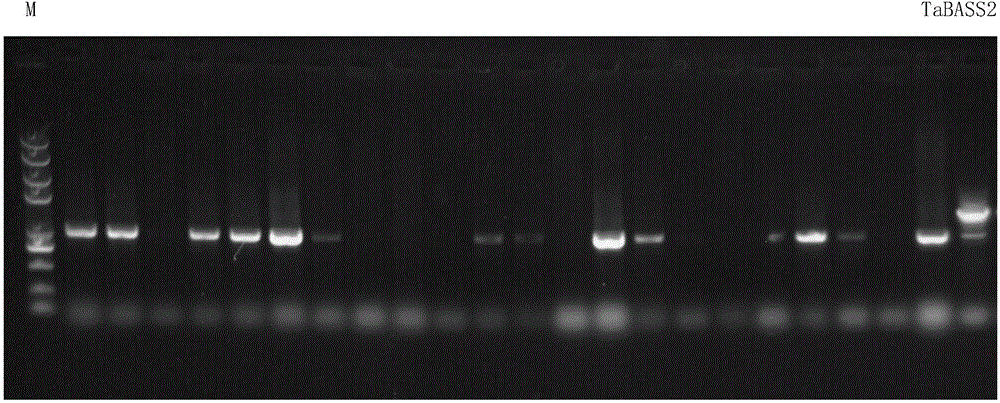
[0065] 2.3 PCR reaction and electrophoresis
[0066] 1) Using cDNA as a template, carry out PCR reaction. Primers are as follows:
[0067] QRT-1: ATAGCGATGACACCACTCCT
[0068] QRT-2: TTTCAACACTTCTGCGACTT
[0069] 2) PCR system:
[0070]
[0071]
[0072] 3) PCR program:
[0073] 95°C for 5min; 25~30cycles 95°C for 30s, 60°C for 30s, 72°C for 30s; 72°C for 5min.
[0074] The number of PCR cycles was determ...
Embodiment 3
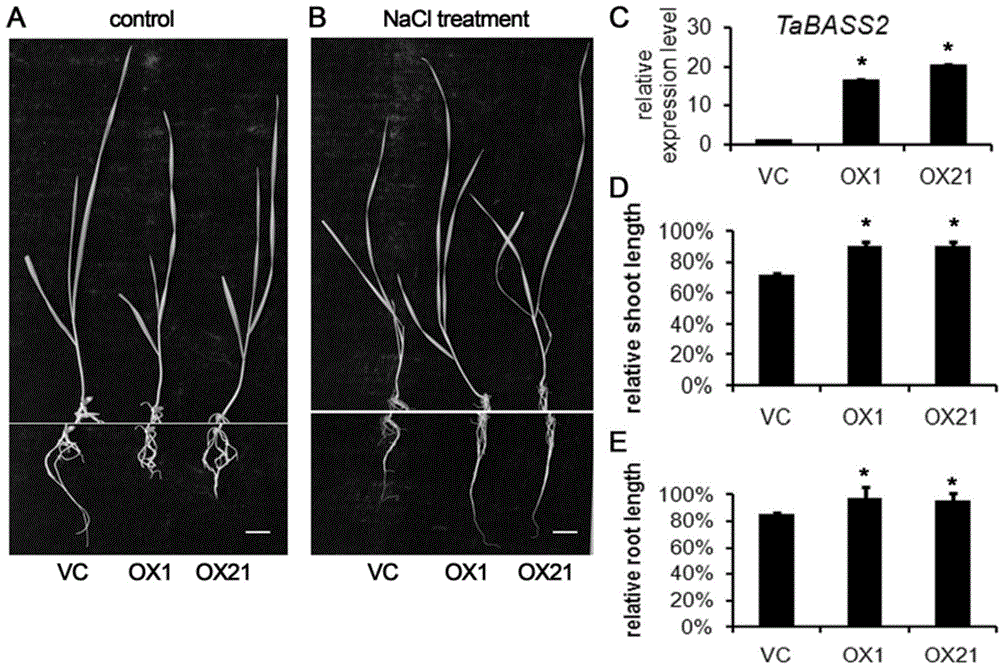
[0076] Embodiment 3, change TaBASS2 wheat salt tolerance analysis
[0077] The plant expression vector pGA626 is an expression vector containing a ubiquitin promoter, and its multiple cloning site contains recognition sites for restriction endonucleases HindIII and BamHI. Based on this, design primers containing HindIII and BamHI recognition sequences upstream of the start codon and downstream of the stop codon of the target gene, use high-fidelity Taq enzyme to amplify the target gene, and use restriction enzymes HindIII and BamHI to amplify the vector pGA626 and the target gene The product fragments were digested separately. The fully digested carrier is separated by electrophoresis on 1% agarose gel, recovered by gel, and connected with the amplified fragment of the digested target gene. Transform Escherichia coli competent, use the primer tttagccctgccttcatacg on the carrier and the downstream primers of the gene for PCR identification, and use HindIII and BamHI for enzyme...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com