A method for improving the efficiency of homologous recombination in Gluconobacter oxidans
A technology of Gluconobacter oxydans and oxidized glucose, which is applied in the field of genetic engineering, can solve the problems of low homologous recombination efficiency of Gluconobacter oxidans, hinder the genome transformation of Gluconobacter oxidans, etc., and achieve the goal of improving the efficiency of homologous recombination and simplifying the construction process Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0016] Example 1 Plasmid pKD46 into G.oxydans WSH-003
[0017] pKD46 was introduced into G. oxydans WSH-003 by electroporation. The specific process is divided into three steps:
[0018] 1. Electrotransfer Competent Preparation:
[0019] 1) Take a G.oxydans WSH-003 glycerol tube and streak it on a sorbitol plate, and incubate at 30°C for 24-48 hours until a single colony can be identified;
[0020] 2) Pick a single colony and insert it into a 250ml Erlenmeyer flask containing 25ml sorbitol liquid medium, pre-cultivate at 30°C, 220rpm for 24-36h, until OD 600 >2;
[0021] 3) Take 2.5ml of the pre-cultured bacterial solution and put it into a 250ml Erlenmeyer flask containing 25ml of sorbitol medium, culture it at 30°C and 220rpm for 6-8h, then measure the OD 600 , to OD 600 to 1.6-1.8;
[0022] 4) Centrifuge at 5000rpm for 5min at 4°C, remove the supernatant;
[0023] 5) Place the cells on ice, add 20ml of 10% glycerol solution pre-cooled to 0°C to resuspend the cells to...
Embodiment 2
[0037] Example 2 Construction of Homologous Fragments
[0038] Take the knockout of the membrane gluconate dehydrogenase gene of Gluconobacter oxidans as an example.
[0039] 1) Construction of screening markers
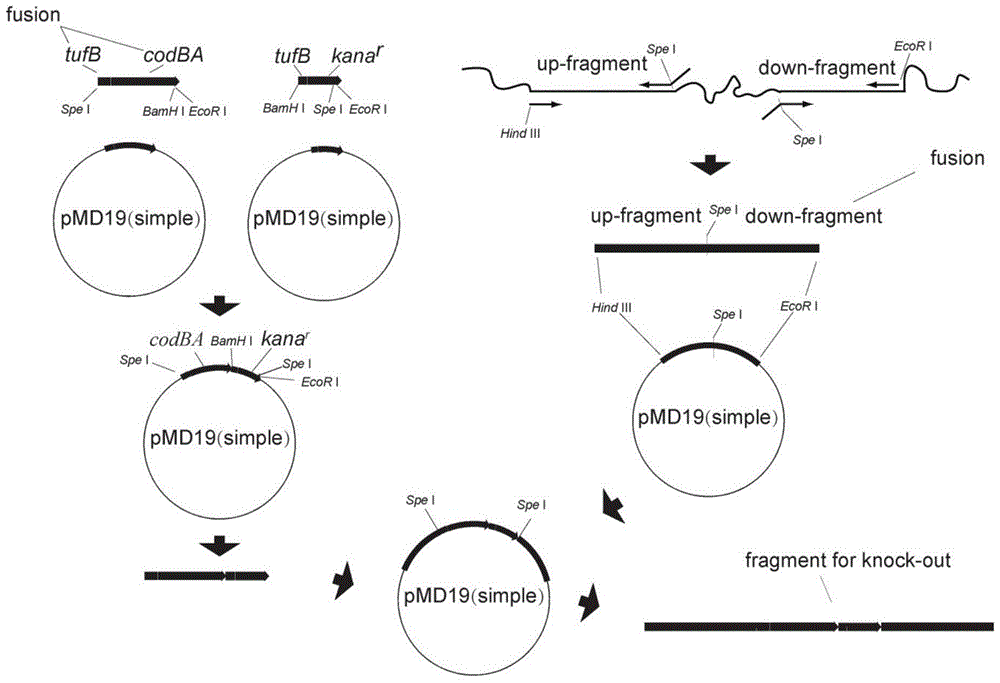
[0040] Primers were designed to amplify the kana resistance gene (kana, see SEQ ID NO. 2 for the sequence) derived from the plasmid pBBRMCS-2, including the open reading frame and the terminator. Primers were designed to amplify the cytosine deaminase gene (codBA, see SEQ ID NO.3) derived from the genome of Escherichia coli, including the open reading frame and the terminator. Primers were designed to amplify the tufB promoter derived from the genome of G. oxydans WSH-003 (see SEQ ID NO.4 for the sequence). The open reading frames of Kana and cytosine deaminase were connected to the promoters by fusion PCR to form two complete marker genes, and the two marker genes were connected by restriction enzyme digestion, and the two marker genes were connected together in p...
Embodiment 3
[0045] Transformation of embodiment 3 knockout fragments and verification of recombinant bacteria
[0046] The knockout fragment was recovered from pMD19 (Simple) constructed by enzyme digestion, and the concentration was about 100 ng / μl. Transformation into G.oxydans WSH-003 / pKD46 was performed according to the transformation method of plasmid pKD46. The difference is: add sorbitol medium containing 10mmol / L arabinose after electric shock; after post-cultivation, spread on a plate containing 10μg / ml Kanna resistance. The grown single colony was verified by the colony PCR method for the marker gene. A positive verification indicates successful recombination.
[0047] When removing the marker, use the above-mentioned recombinant bacteria G.oxydans WSH-003 / pKD46 to prepare electroporation competence, and recover the fused homology arm fragment without the marker gene from the constructed T vector, and electroporate the fragment, the method is the same as the knockout fragment ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com