Carbonyl reductase gene, codase, vector, engineering bacterium and application thereof
A technology of genetically engineered bacteria and reductase, applied in genetic engineering, oxidoreductase, application, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1: Amplification of the carbonyl reductase gene adh5
[0041] According to the whole genome sequencing information of Burkholderia gladioli (Burkholderia gladioli) ZJB-12126, a large number of carbonyl reductases were excavated, one of which has the ability to catalyze 2-benzamidomethyl-3-ketobutyrate, (R)-tert-butyl 6-cyano-5-hydroxy-3-carbonylhexanoate and ethyl 4,4,4-trifluoroacetoacetate generate (2S,3R)-2-benzamidomethyl- 3-Hydroxybutyrate, tert-butyl 6-cyano-(3R,5R)-dihydroxyhexanoate, and (S)-4,4,4-trifluoro-3-hydroxybutyrate-functional enzymes are The carbonyl reductase BgADH5 involved in the present invention.
[0042] Using FastDNA from MPBio The Spin kit was used to extract the total genomic DNA of Burkholderia gladioli ZJB12126 cells. Using the genomic DNA as a template, primer 1 (5'-ATGGCAGACGTCAACAGCCTGTTC-3'), primer 2 (5' -TCAGACCGTGCTGGTGAGGCC-3') for PCR amplification. PCR reaction system (total volume 50 μL): 5 μL of 10×Pfu DNA Polymerase...
Embodiment 2
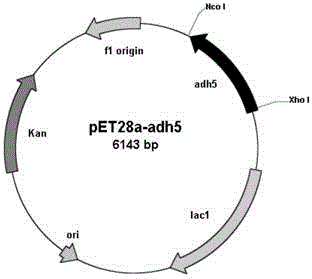
[0045] Embodiment 2: Construction of recombinant Escherichia coli BL21(DE3) / pET28a-adh5
[0046] Design primer 3 (5'- CCATGG CAGACGTCAACAGCCTGTTC-3'), primer 4 (5'- CTCGAG GAC
[0047] CGTGCTGGTGAGGCC-3'), and NcoI and XhoI restriction enzyme sites (underlined) were introduced into primer 3 and primer 4, respectively. Under the priming of primer 3 and primer 4, high-fidelity PfuDNA polymerase was used to amplify to obtain a 774bp carbonyl reductase gene sequence (its nucleotide sequence is shown in SEQ ID NO: 1), and after sequencing, Nco The amplified fragment was treated with I and Xho I restriction enzymes (TaKaRa) and ligated with the commercial vector pET28a (Invitrogen) treated with the same restriction enzymes using T4 DNA ligase (TaKaRa) , construct the expression vector pET28a-adh5. The constructed expression vector pET28a-adh5 was transformed into Escherichia coli BL21(DE3) (Invitrogen), spread on LB plates containing kanamycin resistance at a final concentratio...
Embodiment 3
[0048] Embodiment 3: Recombinant carbonyl reductase (BgADH5) wet thallus
[0049] The recombinant Escherichia coli E.coliBL21(DE3) / pET28a-adh5 bacterial cell containing the expression recombinant plasmid pET28a-adh5 obtained in Example 2 was inoculated into LB liquid medium containing a final concentration of 50 μg / mL kanamycin resistance, 37 Cultivate at 200rpm for 12h, then inoculate with 1% inoculum (v / v) into fresh LB liquid medium containing kanamycin resistance at a final concentration of 50μg / ml, and cultivate at 37°C at 150rpm until the bacteria Body OD 600 After reaching 0.6-0.8, add IPTG with a final concentration of 0.1mM, induce culture at 28°C for 12h, centrifuge at 5000rpm at 4°C for 5min, discard the supernatant, collect the precipitate, and obtain the recombinant Escherichia coli BL21( DE3) / pET28a-adh5 wet cells. The bacterium can be used directly as a biocatalyst or for protein purification.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com