Leucine dehydrogenase as well as preparation method and application thereof
A leucine dehydrogenase and amino acid technology, applied in the fields of botany equipment and methods, biochemical equipment and methods, applications, etc., to achieve the effects of reducing production costs, prolonging half-life, and improving production efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment l
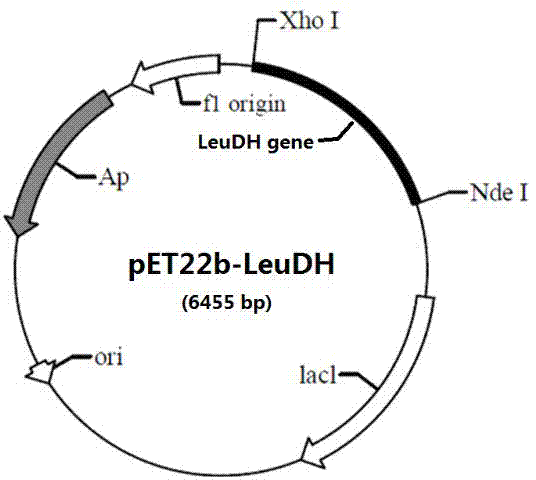
[0041] Embodiment 1: Containing the construction of the recombinant plasmid pET22b-LeuDH of leucine dehydrogenase gene sequence
[0042] Using the database of the National Center for Biotechnology Information (NCBI), we analyzed, screened, and found that there is a sequence in Laceyella sacchari that has a high similarity to the sequence of the leucine dehydrogenase gene, which is likely to encode the leucine dehydrogenase The gene sequence is screened and optimized (optimization includes mutation and insertion of some sites) to obtain the nucleotide sequence with SEQ ID NO.2.
[0043] The Ls-LeuDH gene was artificially synthesized by GenScript according to the nucleotide sequence of SEQ ID NO.2. Add Nde I (CAT) and Xho I (CTCGAG) at both ends of the sequence, respectively, and connect them between the Nde I and Xho I restriction sites on the pET22b plasmid vector. Get the recombinant plasmid pET22b-LeuDH, transfer it into 50 microliters of Escherichia coli competent c...
Embodiment 2
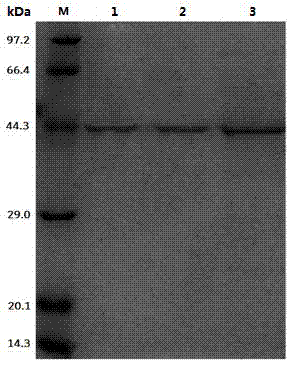
[0045] Example 2: Expression and purification of recombinant leucine dehydrogenase
[0046] Pick engineering bacteria with a single colony E. coli BL21-pET22b-LeuDH, cultured in 10ml LB liquid medium containing 100μg / mL ampicillin with shaking at 37°C for 10 hours; pipette the culture solution according to the inoculum size of 1v / v% and transferred to Cultured at 30°C in a self-inducing liquid medium (the formulation of the self-inducing medium is (per 100 ml): 1.5 grams of peptone, 2.5 grams of yeast powder, 1 gram of sodium chloride, 0.2 grams of glucose, and 0.3 grams of lactose.), to be OD 600nm After reaching 1.0, incubate at 25°C for 20 hours at low temperature, collect the bacterial solution by centrifugation, discard the supernatant medium, wash with equal volume of PBS buffer with pH=8 for three times, add PBS buffer to make the resuspended bacterial solution OD 600nm When it reaches 20, mix well; break with an ultrasonic breaker, ultrasonic working conditio...
Embodiment 3
[0047] Example 3: Determination of Leucine Dehydrogenase Enzyme Activity.
[0048] Each component was added according to the reaction system shown in Table 1, and the expression product of Escherichia coli containing plasmid pET22b was used as a control. The basic principle is that in view of the fact that NADH has a maximum absorption peak at 340nm, the content of the enzyme is quantitatively determined by the change of the light absorption peak. Add in sequence to the cuvette
[0049] Reaction solution 1.95ml: Na at pH 10.0 2 CO 3 / NaHCO 3 Buffer with 10mM L-Leucine, 3.5mM NAD + . Place it in the sample tank of a spectrophotometer, detect the original NADH absorbance value, and immediately add 50 μL of enzyme solution diluted with an appropriate multiple after the reading is stable, start the photometer reading, measure the absorbance value every 1 s, for a total of 30 s, and get △ A 340nm / min value. The enzyme activity unit adopts the international unit IU, that...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com