Method for detecting rumen microorganisms of Holstein cows
A technology of rumen microorganisms and microorganisms, which is applied in the field of detection of animal stomach microorganisms, can solve the problems of long sequencing time, complicated operation, and inaccurate measurement results, and achieve the effect of accurate and comprehensive detection results and reduced detection time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
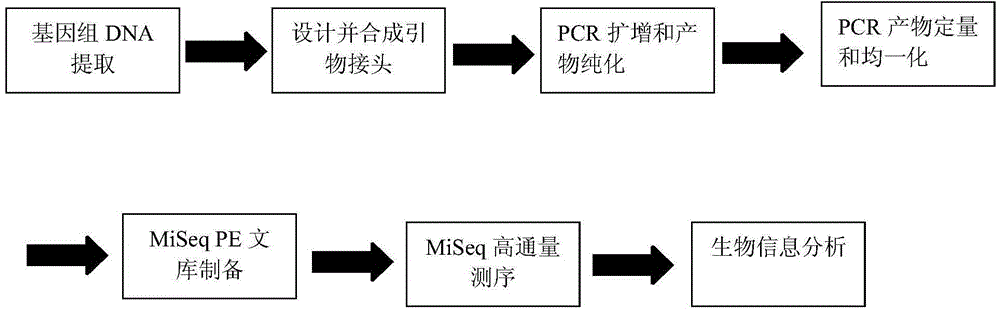
[0031] Embodiment 1: the method of the present invention is: extract the rumen fluid of Holstein milk cow, extract the microorganism DNA wherein and determine its microorganism type and relative quantity by sequencing; Prepare according to the following operation steps:
[0032] (1) Genomic DNA extraction: extract the rumen fluid of Holstein dairy cows, then extract and purify the microbial DNA group, and use 1% agarose gel electrophoresis to detect the extracted genomic DNA;
[0033] (2) Polymerase chain reaction: Synthesize specific primers with barcodes according to the designated sequencing region; mix the PCR products of the same sample and detect them by 2% agarose gel electrophoresis, and use the genomic DNA mini kit for gel recovery The PCR product was recovered by cutting the gel of the kit, eluted with trisaminomethane, and detected by 2% agarose electrophoresis; all samples were carried out according to the formal experimental conditions, and each sample was repeated...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com