Test kit for coxsackie virus A6 nucleic acid and test method
A technology for coxsackie virus and detection kits, which is applied in the directions of microorganism-based methods, microbial determination/inspection, biochemical equipment and methods, etc., can solve the problems of waste, high cost, reduce the probability of pollution and low requirements , highly specific and sensitive effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
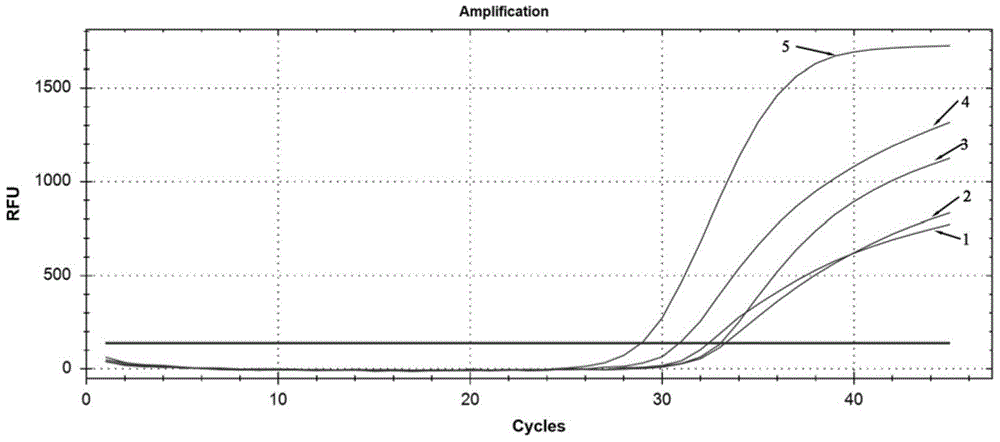
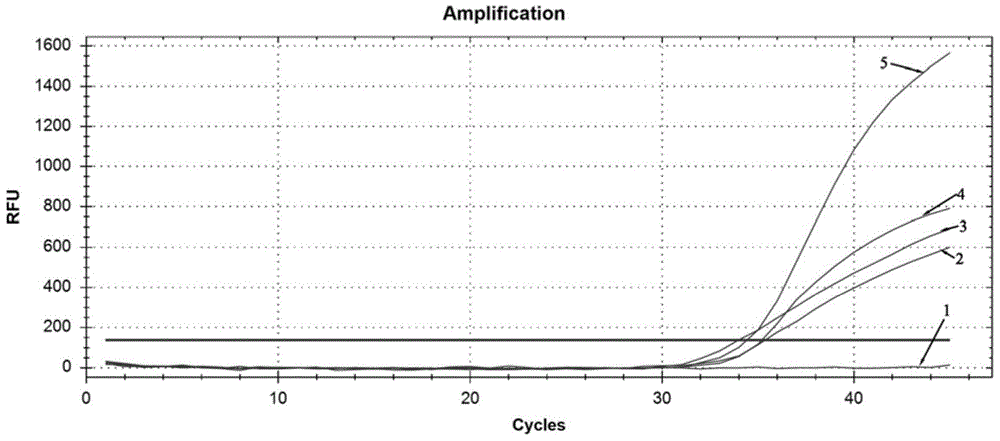
[0056] The present invention searches for the VP1 gene sequence of the Coxsackievirus A6 strain in the Genebank database for the Coxsackievirus A6 type, uses DNAMAN 7.0 software to compare multiple sequences, and finds out the conserved segment. Use GenScript and Oligo 7.0 software to design specific primers and Taqman probes in this segment, and perform BLAST comparison on the NCBI database, and design five sets of primers and probes respectively. The sequences of primers and probes are shown in Table 1 Show. Two CVA6 clinical positive samples were respectively amplified by PCR, and the amplification curves were as follows: figure 1 and figure 2 .
[0057] Table 1 Primer and Probe Sequences
[0058]
[0059] Depend on figure 1 and figure 2 The results show that the amplification curves of the fifth group of primers and probes are the most typical, with obvious exponential phase and plateau phase, higher fluorescence intensity (ordinate value), and smaller CT value (...
Embodiment 2
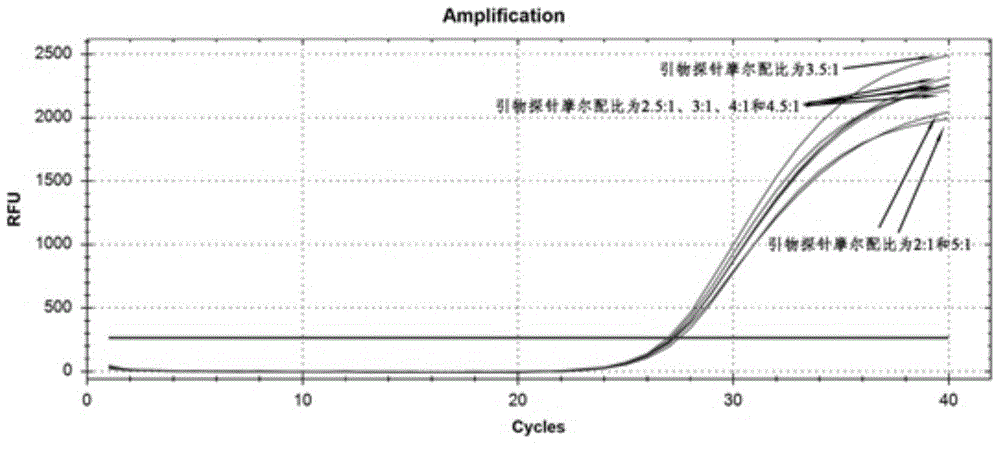
[0061] For the fifth set of primers and probes, the molar ratio of "primer:probe" is 5:1, 4.5:1, 4:1, 3.5:1, 3:1, 2.5:1 and 2:1 , to test the CVA6 clinically positive samples to investigate the effect of different molar ratios of "primer:probe" on the amplification curve. The results are shown in Table 2 and image 3 .
[0062] Table 2 The effect of the molar ratio of "primer:probe" on the amplification curve
[0063] Primer probe ratio
[0064] CT value
[0065] From Table 2 and image 3 It can be seen from the results that the results of the above-mentioned ratio experiments are all good, and can be interpreted normally. However, when the molar ratio of primer to probe is 3.5:1, the curve is the best, and the curve rises higher (ordinate) than the other six ratios, and the CT is relatively smaller (abscissa), indicating that the fluorescence intensity is large The exponential phase of the response comes earlier and the curve rises earlier.
Embodiment 3
[0066] Embodiment 3: the nucleic acid detection kit of Coxsackievirus A6 type of the present invention
[0067] The kit of the invention comprises a real-time fluorescent PCR reaction solution, a reverse transcription system, a primer mixture, a specific fluorescent probe, a Coxsackie virus A6 standard product and DEPC treated water.
[0068] The real-time fluorescence PCR reaction liquid of the present invention comprises 2×PCR buffer, heat-resistant Taq DNA polymerase, magnesium sulfate, and deoxyribonucleotide triphosphate mixture.
[0069] The reverse transcriptase in the reverse transcription system of the present invention is MMLV reverse transcriptase.
[0070]In the primer mixture of the present invention, the base sequence of the forward primer is shown in SEQ ID No.1, the base sequence of the reverse primer is shown in SEQ ID No.2, the forward primer and reverse primer The molar ratio of the primers is 1:1 for SEQ ID No.1:SEQ ID No.2.
[0071] The base sequence of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com