Method, primer, probe and kit for detecting cronobacter sakazakii
A Sakazaki Cronor rod and probe technology, which is applied in biochemical equipment and methods, microbial determination/inspection, resistance to vector-borne diseases, etc. It is unable to adapt to the specific detection of Cronobacter, and nucleic acid detection methods have not been reported, so as to avoid subjectivity, have a wide range of applications, and improve specificity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
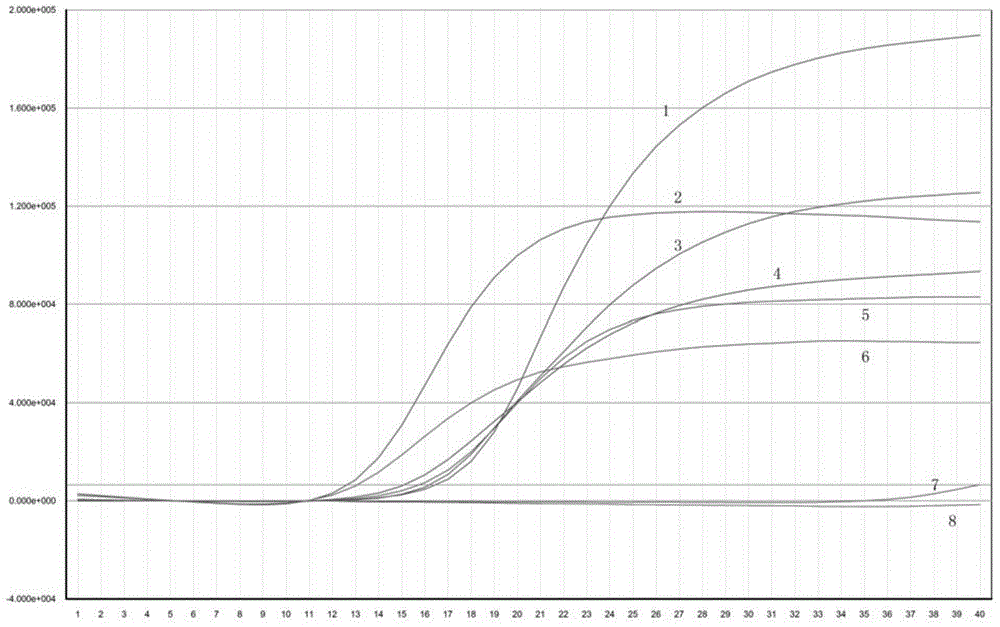
Image
Examples
Embodiment 1
[0043] A method for detecting Cronobacter sakazakii, carried out according to the following steps:
[0044] 1. Design and synthesis of primers and probes
[0045] Download all genome sequences of Cronobacter sakazakii from the NCBI database, and select a highly conserved segment (SEQ ID NO: 1) with no secondary structure through comparative analysis of the sequences. The sequence is as follows: CGCTGTACATTCGCCCCTTTGATGAAAGCGTGCTGAATAAAGATCTCTCAGACAGCCCGCTTTTTAC CGCGCCAGGGCAGCGCCACCTGGCTGTCGGCGC TTGCGCGCTCGACGCGTTACCCGATTGTCGTGGTGGC GCGCGACCTGGCTTAAAACAGTATTCCCGATTTGCTTATCAACGGTGCGCTGCTGACTGCCATTATCCTGATGGGCA
[0046] Among them, bold italics are the upstream and downstream primer sequences, and bold underlines are the probe sequences.
[0047] Primers and Taqman probes were designed for the above segments using the biological software Primer Express 3.0. And the Blast homology comparison verification was carried out through the NCBI database to ensure that the sequence ...
Embodiment 2
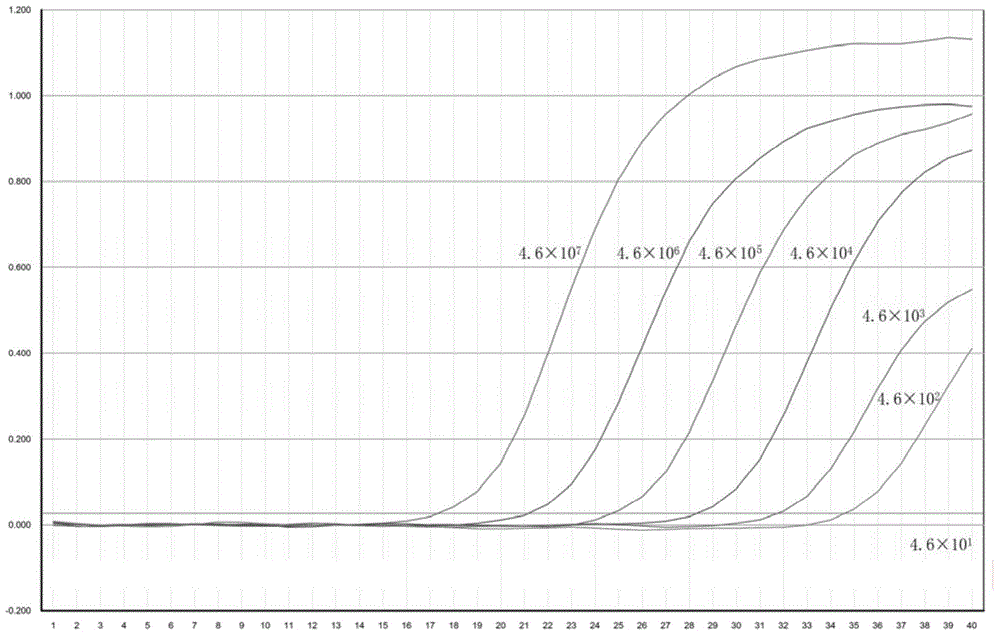
[0076] The extracted Kronobacter sakazakii ATCC 29544 DNA was diluted with DEPC (diethylpyrocarbonate)-treated water gradient to 4.6×10 7 , 4.6×10 6 , 4.6×10 5 , 4.6×10 4 , 4.6×10 3 , 4.6×10 2 , 4.6×10 1 Copy number / mL, utilize the fluorescent quantitative PCR reaction system that establishs in embodiment 1 to carry out relative sensitivity detection, detection result is as follows figure 2 shown.
[0077] The results confirmed that the detection limit of Cronobacter sakazakii reached 460 template copies / ml.
Embodiment 3
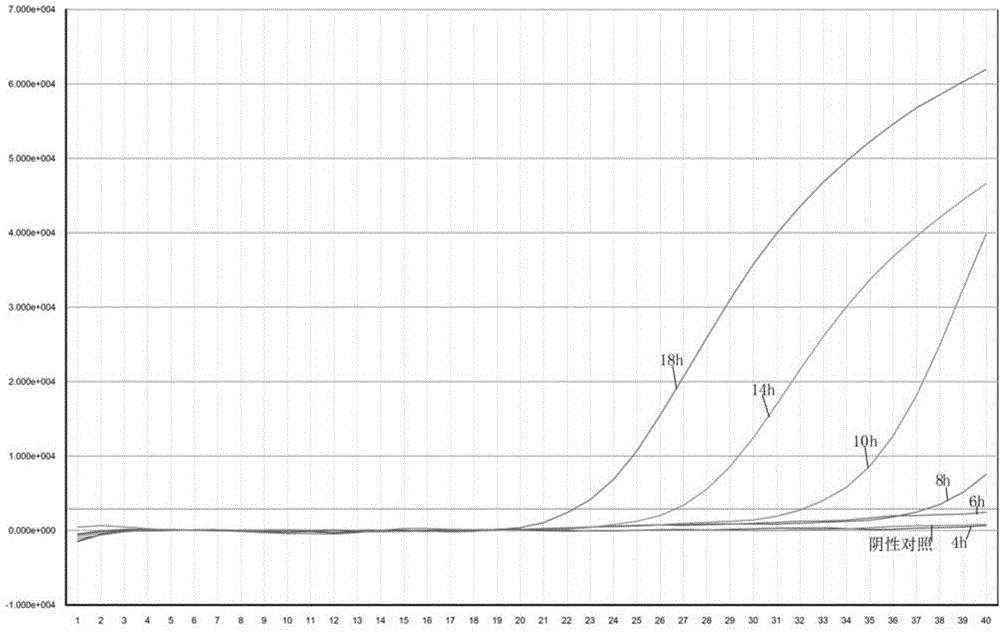
[0079]Infant formula milk powder was taken, and it was tested to be free of Cronobacter sakazakii contamination according to the GB 4789.40-2010 method. Take 1ml of bacterial suspension with a concentration of 10cfu / ml and mix it evenly in 25g of infant formula milk powder, let it stand for 10min, add 225ml of BPW (buffered peptone water) bacterial enrichment solution, mix evenly, culture at 37°C, and incubate at 4h, 6h, After 12h, 18h, and 24h, take 1ml of enrichment solution, centrifuge at 4000r / min for 10min, carefully pick out the upper layer of fat, centrifuge at 12000r / min for 5min, discard the supernatant, and extract DNA as shown in Example 1. Fluorescent PCR detection was performed according to the PCR reaction conditions established in Example 1, and DEPC water was used as a negative control.
[0080] The result is as image 3 As shown, using fluorescent quantitative PCR to detect artificially contaminated infant formula milk powder with a dose of 10cfu / 25g homogena...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com