Microbiology method for detecting metallic arsenic in water body
A technology for metal arsenic and water body is applied in the field of microbiology for detecting metal arsenic in water body, which can solve the problems of slow development of heavy metal microbial detection technology, and achieve the effects of easy promotion, reliable results and stable detection signals.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Embodiment 1. Preparation of ΔarsB mutant bacteria
[0066] 1.1 Primer information and synthesis
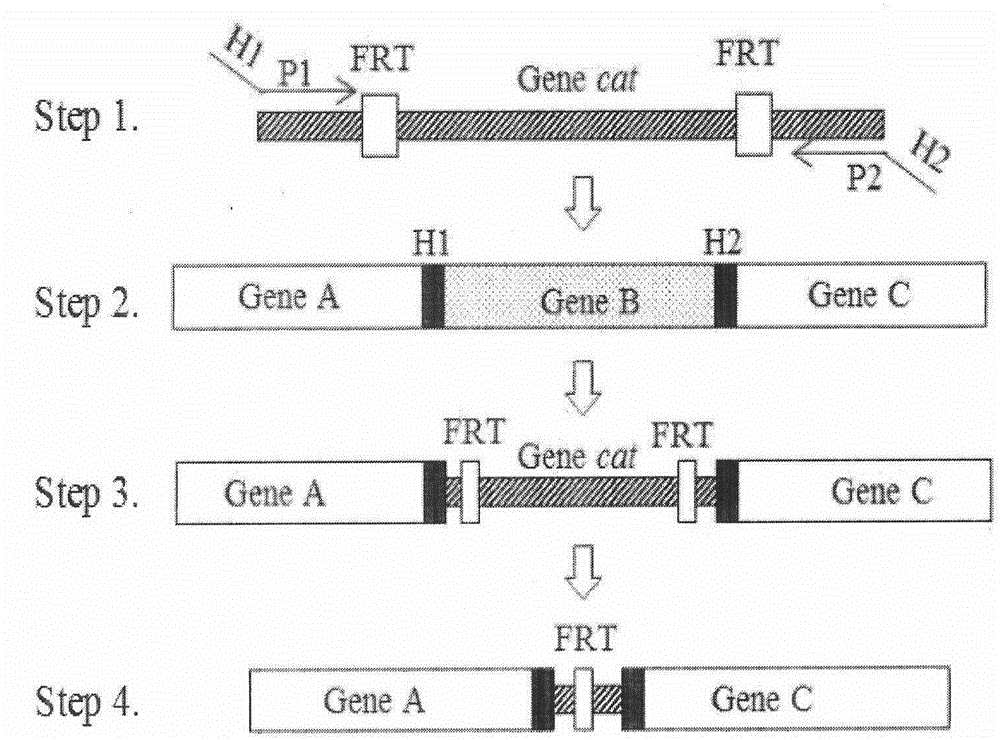
[0067] Knockout Primers: According to http: / / ecogene.org / Provide primer sequences for gene knockout, Primer5.0 and DNAMAN software, design homologous recombination primers (see Table 1), the 5' end is the homology arms on both sides of the arsB gene, and the 3' end is used to amplify chloramphenicol resistance gene. upstream homology arm primer H 1 -P 1 ; Downstream homology arm primer H 2 -P 2 .
[0068] Knockout Identification Primers: Using the Harvard Molecular Technology Group & Lipper Center for Computational Genetics website
[0069] http: / / arep.med.harvard.edu / labgc / adnan / projects / EcoliKO-primers / EcoliKOprimers.htm The provided pair of knockout identification primers designed spanning the outside of the gene to be knocked out (see Table 2). Identification of arsB gene knockout primer design: the upstream primer arsB-jianding-F is located 108bp upstream ...
Embodiment 2
[0102] Embodiment 2. Construction of fusion reporter gene pars-arsR-gfpmut2
[0103] 2.1 Primer information and synthesis
[0104] According to the target gene pars and arsR sequence published by GenBank and the known gfpmut2 gene sequence, use the Primer5.0 software to design primers (see Table 5), and some primers introduce restriction sites at the 5' end or 3' end of the gene. Pars-arsR gene 5′ end primer (P 3 : pars-arsR-F) and 3' end primer (P 4 :pars-arsR-R), make P 4 with P 5 Complementary sequences exist between, P s (gfpmut2-F) and P 6 (gfpmut2-R) is a pair of primers for amplifying the gfpmut2 gene, and M13F and M13R are primers for identifying the pars-arsR-gfpmut2 fragment on the pMD19-T vector. Design the inner primers (P 7 : araB-Ni and P 8 : araB-Ci) and outer primer (P 9 :araB-No and P 10 : araB-Co), gene knockout two outer primers unchanged, P 7 with P 8 A pair of gene primers for amplifying pars-arsR-gfpmut2, P 11 with P 12 A pair of gene primer...
Embodiment 3
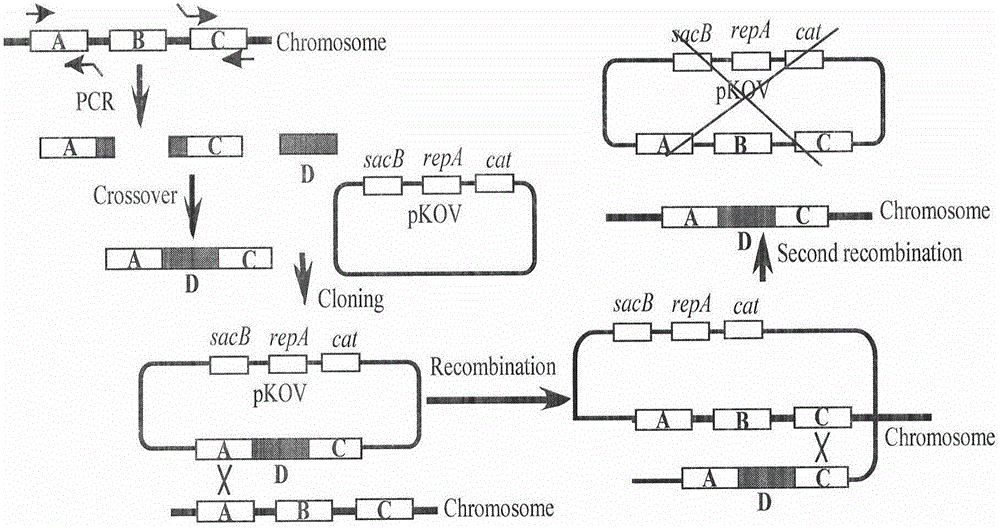
[0140] Example 3 Gene knock-in
[0141] 3.1 Fusion of gene knock-in fragments
[0142] 3.1.1 Amplification of two homology arms:
[0143] Take 1mL of the overnight bacterial solution of E.coli MC4100 in a 1.5mL EP tube, centrifuge at 8000rpm for 3min, discard the supernatant, and use 1mL MilliQ H 2 After O was resuspended and washed twice, boiled in water for 10 min, centrifuged at 12 000 rpm for 3 min, and 24 μL of the supernatant was taken as a template. According to the table dubbed system, do PCR identification.
[0144]
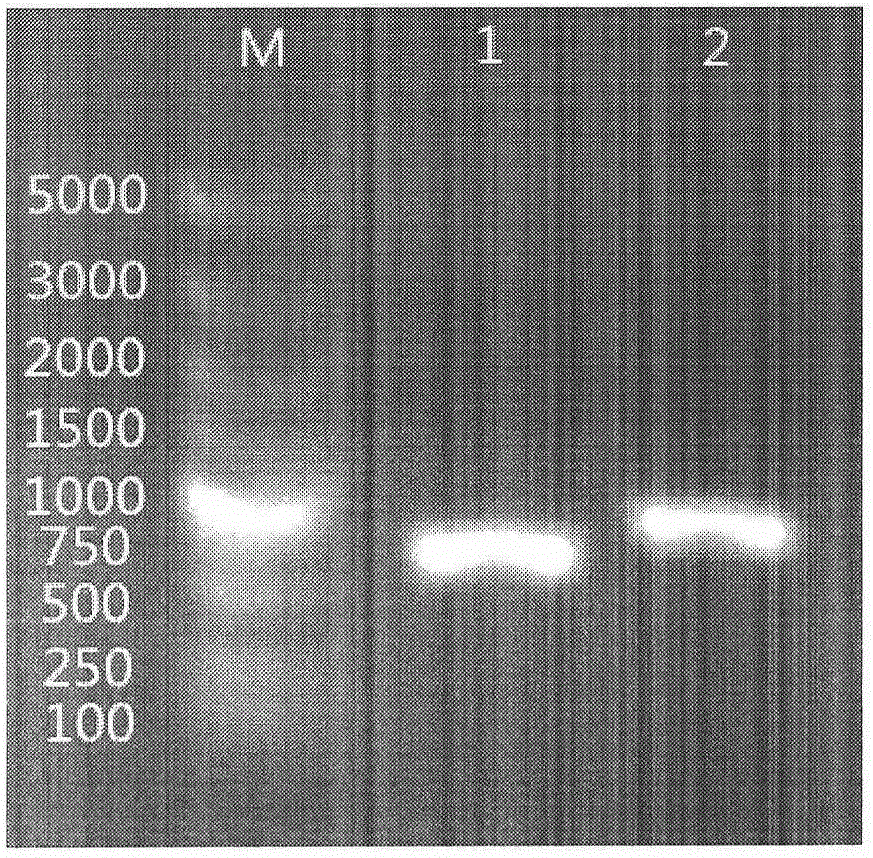
[0145] The PCR reaction conditions were as follows: 94°C, 1min; 88°C, 4min; 94°C, 10sec, 66°C, 3min, 25 cycles; 72°C, 10min, 4°C storage. 5 μL of the product was identified by 1.0% Agarose gel electrophoresis. Purify the PCR product with a purification kit, and finally add an appropriate amount of sterile MilliQ H 2 O dissolved and eluted for later use.
[0146] 3.1.2 Amplification of the pars-arsR-gfpmut2 gene:
[0147] Take 1 mL of the overni...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com