Phyllachora dalbergiicola molecular detection primer and fast detecting method for dalbergia odorifera
A Dalbergia Dalbergia, molecular detection technology, applied in biochemical equipment and methods, microbial measurement/inspection, DNA/RNA fragments, etc., can solve the problems of isolating pathogenic bacteria, time-consuming, heavy workload, etc., and achieve high sensitivity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
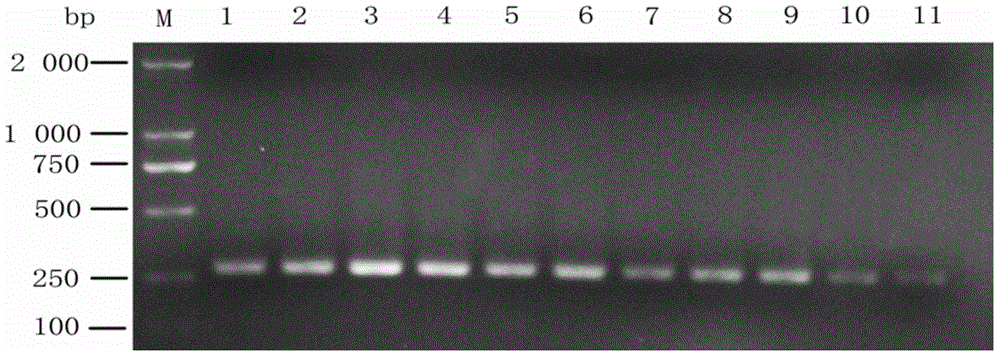
[0055] Embodiment 1: Detect the specific amplification of the specific primer designed by the present invention to Dalbergia nigra
[0056] 1. Preparation of Black Mole Leaf Samples
[0057] See the test samples listed in Table 1 and their collection locations. After the diseased leaf samples of Dalbergia sinensis, white grass, Pennisetum, Goose-view grass and Gangzhu black nevus were surface-sterilized with 75% alcohol, the diseased leaves were cut into 4 × 4 mm tissue blocks using sterilized scissors. Grind with liquid nitrogen. Sample numbers of black mole diseased leaves: B01~B13.
[0058] Table 1 Collection sites and molecular detection results of each tested sample
[0059]
[0060] "+" indicates that the target band of 273 bp can be detected, and "-" indicates that the target band of 273 bp cannot be detected.
[0061] 2. Preparation of Control Strain Samples
[0062] (1) Use sterilized scissors to cut the diseased and healthy junctions of typical lesions, and c...
Embodiment 2
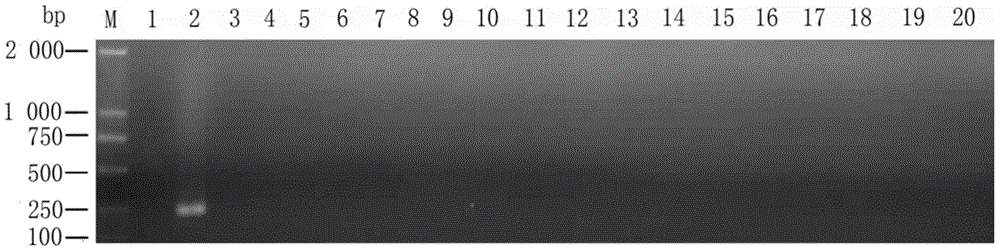
[0072] Example 2: Sensitivity detection of primers to Dalbergia nigra
[0073] 1. DNA concentration dilution
[0074] After the concentration of the extracted Dalbergia nigra genomic DNA was determined by ultraviolet spectrophotometer, the Dalbergia nigra genomic DNA was gradually diluted down to 10ag / μL by a 10-fold order of magnitude starting from a concentration of 100ng / μL.
[0075] 2. Sensitivity detection of specific primers
[0076] The first round of PCR reaction: ITS4 / ITS5 was used as the primer to carry out the first round of PCR, and the amplification reaction system was: 2×Taq PCR MasterMix 12.5 μL, step 1) The total DNA of Dalbergia officinale leaves was 1 μL, 10 μmol / L ITS4 1 μL each of / ITS5, with ddH at the end 2 O supplemented to 25 μL, centrifuged for 15 sec and placed in a PCR instrument for amplification; PCR reaction conditions were: 94 °C pre-denaturation for 4 min; 94 °C denaturation for 30 sec, 55 °C annealing for 30 sec, 72 °C extension for 1 min, a ...
Embodiment 3
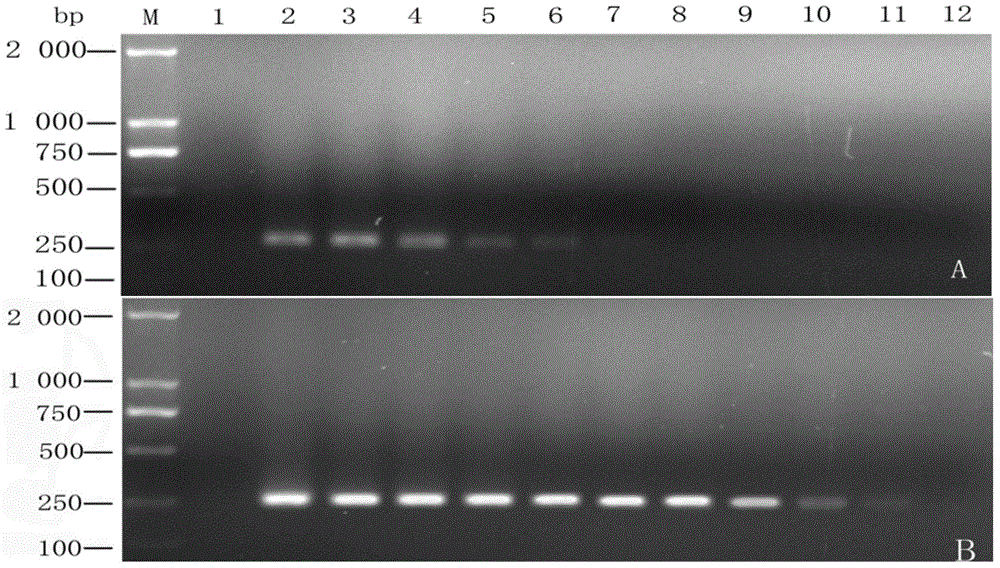
[0081] Embodiment 3: the detection of Dalbergia nigra bacteria in the diseased tissue of Dalbergia sinensis
[0082] 1. Sample Collection
[0083]A total of 23 samples of diseased and healthy tissues of Dalbergia japonica leaves were collected from the state-owned Chengmai Forest Farm in Hainan Province, and were detected by nested PCR.
[0084] 2. DNA extraction and detection
[0085] Using the plant tissue DNA extraction method described in step 2 in the summary of the invention, carry out nested PCR amplification according to the above-mentioned implementation method, PCR reaction system 25 μ L, nested PCR reaction system uses primer ITS4 / ITS5 as the first round of reaction primers, take 1 μL of the first-round PCR reaction product was used as a template, combined with Dalbergia spp. specific primers for the second-round PCR amplification, and the reaction procedure and detection method were the same as those described in the summary of the invention.
[0086] 3. Test res...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com