Visual chip and preparation method thereof and method for chip visualization
A technology of chips and biochips, applied in the field of biochips and chip visualization, can solve problems such as complexity, inconvenient interpretation of results, error-prone, etc., and achieve the effect of simple operation, convenient observation and reading, and less error-prone
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] The preparation of embodiment 1 chip
[0043] 1. Procurement of probes and equipment: All probes were purchased from Shanghai Sangon Biotechnology Co., Ltd., and the spotting instrument was Jingxin PersonalArrayer 16 personal spotting instrument, which was purchased from Boao Biological Co., Ltd.
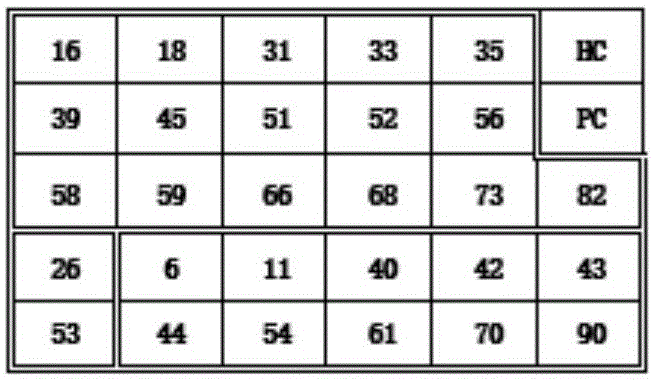
[0044] 2. Setting of the probe style on the chip: according to the project requirements, set the corresponding program on the spotting instrument software, that is, set the spotting style of the probe to be spotting as figure 1 The numbers and letters shown in .
[0045] 3. Spotting: place the film to be spotted on the film placement place of the spotting instrument, and spot the sample according to the settings, such as figure 1 , and carry out drying treatment. That is, the corresponding chip is obtained.
[0046] Spotting of the probe: use the setting program of the spotting instrument to spot the probe solution on the chip with the name of the corresponding detection i...
Embodiment 2
[0047] The detection of embodiment 2 human papillomavirus (HPV) nucleic acid typing (28 types)
[0048] 1. Purchase of HPV nucleic acid typing primers and probes: including two sets of primers and 30 sets of specific probes, of which two sets of primers are used The other set of probes is hybridization reaction quality control probes, as long as the hybridization process is normal, it can be displayed in the end; the remaining 28 sets of probes are used for typing of 28 specific types , were purchased from Shanghai Sangon Biotech.
[0049] 2. Extraction of HPV nucleic acid samples, the HPV nucleic acid samples are extracted from cervical exfoliated cell specimens of HPV patients in a hospital in Jiangsu Province.
[0050] 3. PCR amplification: Prepare a PCR system according to the reagents and nucleic acid samples in steps 1 and 2, a total of 20ul system, including 19ul of PCR reaction solution and 1ul of template. Carry out PCR amplification according to the following proce...
Embodiment 3
[0059] Preparation and detection of embodiment 3 protein chip
[0060] 1. Preparation of the protein chip: immobilize the capture antibody on the solid phase support in the form of the pattern to be displayed (including numbers and letters). The solid phase support can be a glass slide or a nitrocellulose membrane.
[0061] 2. Sample treatment: label the sample with protein biotin, and covalently couple the activated biotin to the amino group of the protein.
[0062] 3. After the biotin-labeled sample is added to the chip and incubated with the reaction, the capture antibody can bind to the specific protein.
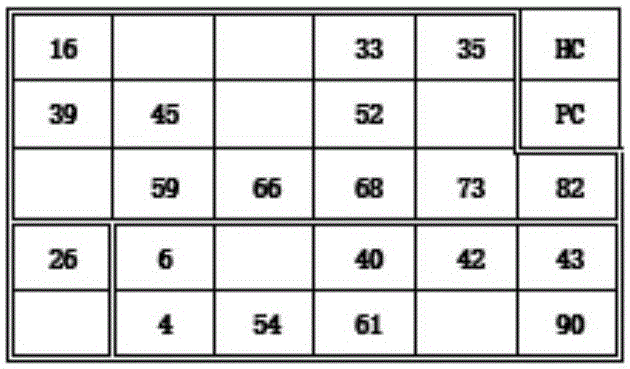
[0063] 4. Add HRP-streptavidin to combine with chemiluminescent substrate or fluorescent dye-streptavidin as signal detection, and observe the corresponding experimental results on the chip after 10 minutes of reaction. Color rendering effect such as image 3 As shown, the test results can be read directly without providing another comparison table.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com