Detection kit and detection method for flavobacterium columnare
A technology of Flavobacterium columnar and detection kit, which is applied in the direction of microorganism-based methods, microbial measurement/inspection, biochemical equipment and methods, etc., can solve the problem that there is no quantitative PCR research report for Flavobacterium columnar, and achieve the protection of fish Healthy, easy-to-use procedures, high sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment l
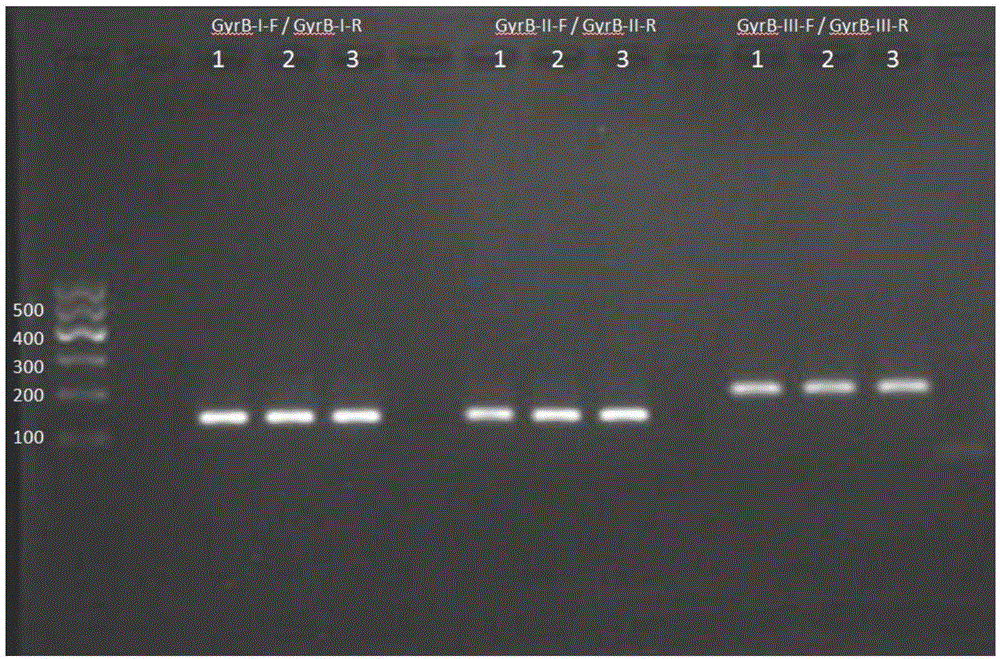
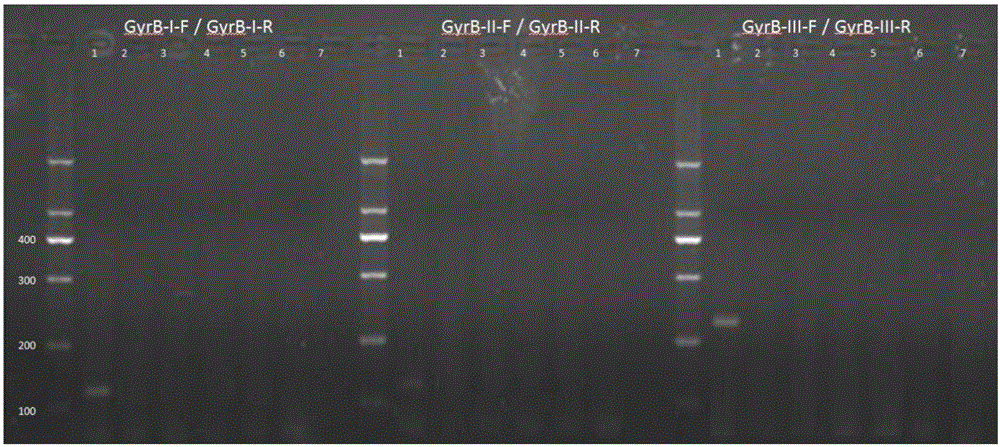
[0028] Example 1: Primer design and screening for fluorescent quantitative PCR detection of Flavobacterium columnar gyrB gene
[0029] 1. Bioinformatics method to design primers and perform primer screening
[0030] Download the full sequence of the gyrB gene of Flavobacterium columnar in GenBank (GeneBank: CP003222.2), and download the full sequence of the gyrB gene of the negative control bacteria. The negative control bacteria include other species of Flavobacterium, Aeromonas, and Pseudomonas See the table below for genera and related species of Vibrio. After comparison by ClustalX, select the appropriate region to design primers. This region is conserved in Flavobacterium columnar species, but there is at least 10%-30% difference compared with the negative control. After selecting the region, use ABIPrimerExpress3.0 real-time fluorescent quantitative PCR Primer design software to design synthetic primers. The primers allow 2 or less degenerate bases at the same variable...
Embodiment 2
[0068] Embodiment 2: Optimization of the amount of fluorescent quantitative PCR primers
[0069] 1. The first optimization of the amount of fluorescent quantitative PCR primers
[0070] With diluted L1 (10 -4 ) and L2 (10 -5 ) The positive control DNA is used as a template for optimizing the amount of primers, and the upstream and downstream amounts of the primers are optimized by serial dilution within the range of 5.0-12.5 pmol. (5U) 0.6uL, real-time fluorescent PCR buffer (2×) 12.5uL (10×buffer2.5uL of Takara Company of 2.5uL, 2uL of 10mMdNTP and the mixture of 10000×SYBRgreenI0.0025uL and 8uLDEPC water are prepared), Make up to 25uL with sterile water. The PCR reaction conditions were as follows: anti-pollution at 37°C for 5 minutes; then pre-denaturation at 95°C for 3 minutes; final amplification at 95°C for 10 sec and 60°C for 40 sec, a total of 40 cycles, and fluorescent signal detection was performed at the end of each cycle of extension. The results are shown in T...
Embodiment 3
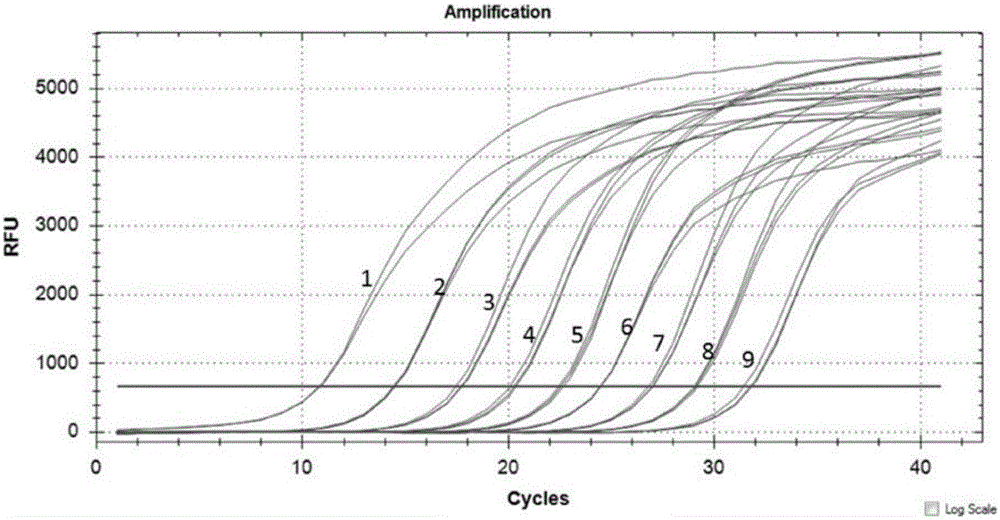
[0081] Example 3: Fluorescent quantitative PCR detection of Flavobacterium columnar gyrB gene
[0082] 1. Establishment of standard curve
[0083] 1.1 Use the positive control plasmid DNA as a template for fluorescent quantitative PCR detection and establish a standard curve. The specific operation is as follows: the plasmid DNA was serially diluted 10 times to I: 1.0×10 10 Copies / uL; II: 1.0×10 9 Copies / uL; III: 1.0×10 8 Copy / uL; Ⅳ: 1.0×10 7 Copy / uL; V: 1.0×10 6 Copy / uL; VI: 1.0x10 5 Copy / uL; VII: 1.0×10 4 Copy / uL; Ⅷ: 1.0×10 3 Copy / uL; IX: 1.0×10 2Copy / uL; X: 1.0×10 1 copy luL;XI: 1.0×10 0 copy / uL. Each dilution was repeated 3 times in parallel. Standard product detection PCR reaction system is: upstream primer 1.5ul (5pmol / ul), downstream primer 2.5ul (5pmol / ul), plasmid DNA 5uL, TaqDNA polymerase (5U) 0.6uL, real-time fluorescent PCR buffer (2×) 12.5 uL (prepared with 10×buffer2.5uL purchased from Takara, 2uL of 10mMdNTP and 10000×SYBRgreenI0.0025uL and 8uLDEPC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com