Wheat 3-hydroxy-3-methylglutaryl-CoA reductase gene tahmgr and its isolation and clone, site-directed mutagenesis and detection method of enzyme function
A technology of methylglutaryl coenzyme and gene site-directed mutation, which is applied in the field of molecular biology and can solve problems that have not been disclosed or published
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
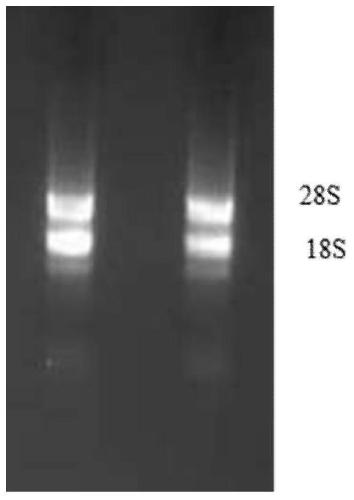
[0078] Example 1 (Wheat variety "Jimai 22" leaf RNA extraction)
[0079] [1] Take about 100mg of wheat leaves and quickly grind them into powder in liquid nitrogen, add 500μl RL (check whether β-mercaptoethanol has been added before use), and immediately vortex vigorously to mix.
[0080] [2] Transfer all the solution to the filter column CS (the filter column CS is placed in the collection tube), centrifuge at 13,000rpm (13,400×g) for 2-5min, carefully pipette the supernatant in the collection tube into the new RNase-Free In the centrifuge tube, the tip should try to avoid contact with the cell debris in the collection tube.
[0081] [3] Slowly add 0.5 times the supernatant volume of absolute ethanol (about 260ul), mix well (precipitation may appear at this time), transfer the obtained solution and precipitation into the adsorption column CR3, 13,000rpm (13,400× g) Centrifuge for 1 min, discard the waste liquid in the collection tube, and put the adsorption column CR3 back i...
Embodiment 2
[0091] Example 2 (synthesis of the first strand of cDNA)
[0092] [1] Add the following reaction mixture to a nuclease-free centrifuge tube in an ice bath:
[0093] 50-500ng mRNA;
[0094] 2 μl oligo(dT)15
[0095] 2μl Super Pure dNTPs (2.5mM each);
[0096] RNase-Free ddH 2 O was adjusted to 14.5 μl.
[0097] [2] After heating at 70°C for 5 minutes, quickly cool on ice for 2 minutes. After brief centrifugation to collect the reaction solution, the following components were added: 4 μl 5×First-Strand Buffer (containing DTT); 0.5 μl RNasin.
[0098] [3] Add 1 μl (200U) TIANScript M-MLV, and mix gently with a pipette.
[0099] [4] Warm bath at 42°C for 50 minutes.
[0100] [5] Heat at 95°C for 5 minutes to terminate the reaction, and place on ice for subsequent experiments or cryopreservation.
[0101] [6] Dilute the reaction system to 50μl with RNase-Free ddH2O, and take 2-5μl for PCR amplification reaction.
Embodiment 3
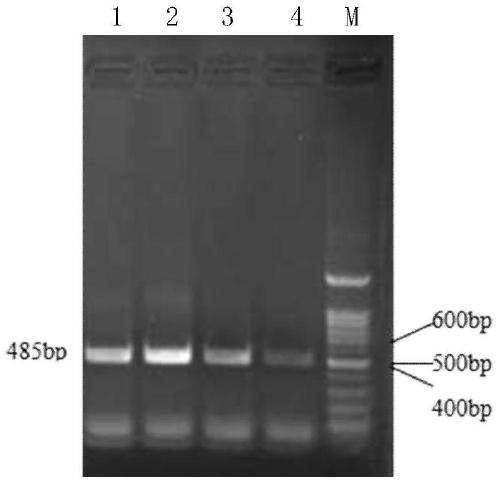
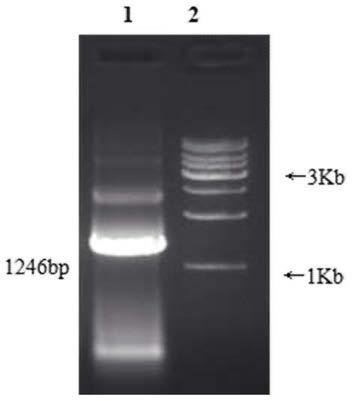
[0102] Example 3 (intermediate sequence amplification, product recovery, connection and transformation, clone identification)
[0103] 1. PCR amplification and detection of the middle sequence of the gene
[0104] By searching and comparing the HMGR sequences of a variety of known plants, especially the plants closely related to wheat, the highly conserved region of the gene sequence was found, and Primer 5.0 was used to design intermediate sequence amplification primers to amplify the corresponding sequence of wheat hmgr.
[0105] Upstream primer mhmgrF1: CGATGGCCGGGAGGAACCTGTACATGAG, the nucleotide sequence of which is shown in Seq ID No.3,
[0106] The downstream primer mhmgrR1:CACCACCAACTGTGCCCACCTCAAT, its nucleotide sequence is shown in Seq ID No.4.
[0107] The PCR system is 50ul, and the components are as follows:
[0108]
[0109] The PCR program was pre-denaturation at 95°C for 5 min, denaturation at 95°C for 30 sec, annealing at 63°C for 30 sec, extension at 72...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com