Electrochemical sensor for detecting kanamycin based on nucleic acid aptamer and preparation method of sensor
A nucleic acid aptamer and kanamycin technology, applied in the field of its preparation, can solve the problems of high cost, low specificity and sensitivity, and achieve the effects of low detection limit, improved sensitivity and high sensitivity detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] The main steps of the electrode modification process are as follows:
[0043] a. The gold electrode is first polished in 0.3 and 0.05 μm alumina slurry until it becomes a mirror surface, and then rinsed repeatedly with PBS and secondary water;
[0044] b. Drop 10 μL of the mixture of HAP2 and Helper (10 μM) onto the electrode surface, and incubate at 37°C for 2 hours. Fix the sulfhydryl chains to the electrode surface through Au-S bonds;
[0045] So far, the modification process of the electrode has come to an end. The following describes the reaction in the homogeneous solution and the main steps in the homogeneous reaction:
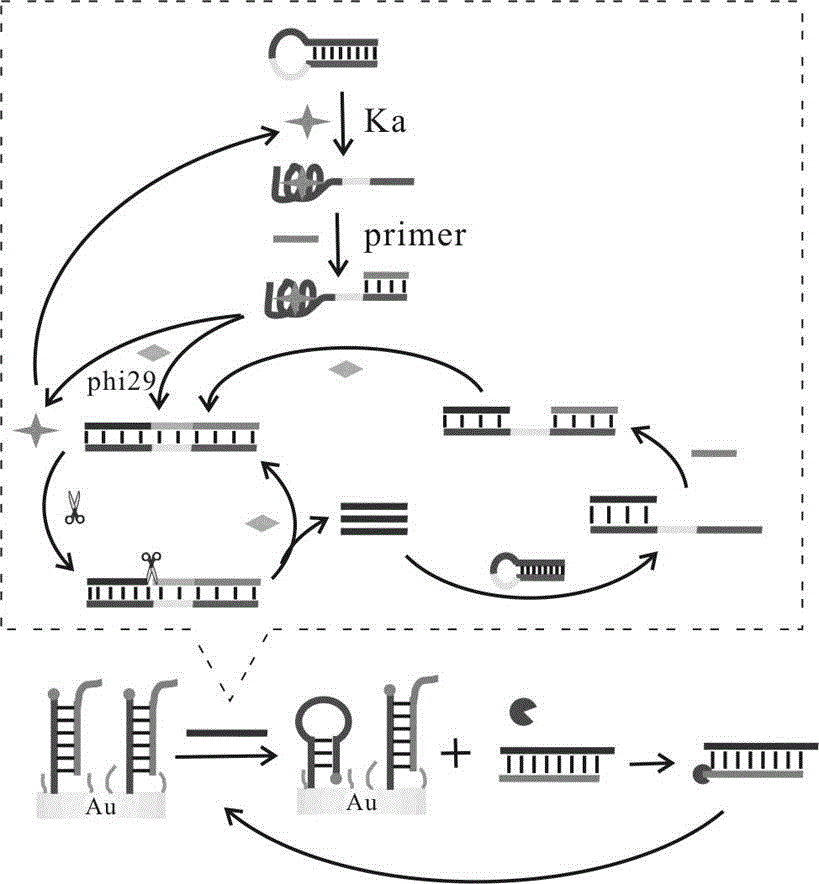
[0046] a. Sterilized water, 10× buffer buffer, different concentrations of HAP1 (1 μM, 2 μM, 5 μM, 10 μM, 15 μM, 20 μM), Primer (10 μM), phi29 DNA polymerase (2 μL), dNTPs (2 μL), Nt.AlwI Endonuclease (1 μL) and the target substance to be tested were added to the centrifuge tube, shaken for 30 seconds, and incubated in a 37°C incubator for 2 ho...
Embodiment 2
[0059] The main steps of the electrode modification process are as follows:
[0060] a. The gold electrode is first polished in 0.3 and 0.05 μm alumina slurry until it becomes a mirror surface, and then rinsed repeatedly with PBS and secondary water;
[0061] b. Drop 10 μL of the mixture of HAP2 and Helper (10 μM) onto the electrode surface, and incubate at 37°C for 2 hours. Fix the sulfhydryl chains to the electrode surface through Au-S bonds;
[0062] So far, the modification process of the electrode has come to an end. The following describes the reaction in the homogeneous solution and the main steps in the homogeneous reaction:
[0063] a. Sterilized water, 10× buffer buffer, HAP1 (10 μM), different concentrations of Primer (1 μM, 2 μM, 5 μM, 10 μM, 15 μM, 20 μM), phi29 DNA polymerase (2 μL), dNTPs (2 μL), Nt.AlwI Endonuclease (1 μL) and the target substance to be tested were added to the centrifuge tube, shaken for 30 seconds, and incubated in a 37°C incubator for 2 ho...
Embodiment 3
[0072] The main steps of the electrode modification process are as follows:
[0073] a. The gold electrode is first polished in 0.3 and 0.05 μm alumina slurry until it becomes a mirror surface, and then rinsed repeatedly with PBS and secondary water;
[0074] b. Add 10 μL of the mixture of HAP2 and Helper at different concentrations (1 μM, 2 μM, 5 μM, 10 μM, 15 μM, 20 μM) onto the electrode surface and incubate at 37°C for 2 hours. Fix the sulfhydryl chains to the electrode surface through Au-S bonds;
[0075] So far, the modification process of the electrode has come to an end. The following describes the reaction in the homogeneous solution and the main steps in the homogeneous reaction:
[0076] a. Sterilized water, 10× buffer, HAP1 (10 μM), Primer (10 μM), phi29 DNA polymerase (2 μL), dNTPs (2 μL), Nt.AlwI Endonuclease (1 μL) and the target substance to be tested were added to the centrifuge tube, shaken for 30 seconds, and incubated in a 37°C incubator for 2 hours.
[...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com